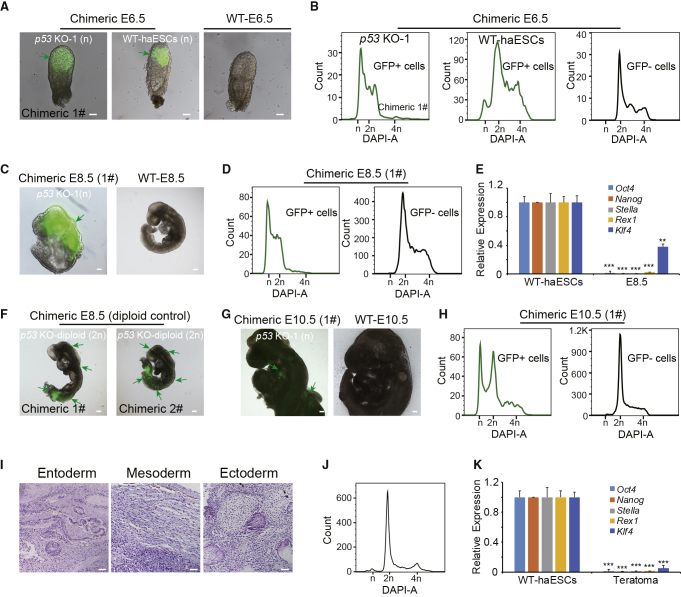
Figure 3.
In Vivo Analysis of p53-KO haESCs
(A) Morphology of chimeric embryos at E6.5 derived from GFP-labeling WT-haESCs and p53 KO-1 cells. Scale bar, 100 μm. WT-E6.5 embryo was used as a control.
(B) DNA-content analysis of GFP+ cells and GFP− cells in chimeric E6.5 embryos derived from GFP-labeling WT-haESCs and p53 KO-1 cells. The percentages of the 1n (G0/G1) peak in GFP+ cells from p53 KO-1 E6.5 and WT-haESCs E6.5 were 28.5% and 4.7%, respectively.
(C) Morphology of chimeric embryo at E8.5 derived from GFP-labeling p53 KO-1 cells. WT-E8.5 embryo was used as a control.
(D) DNA-content analysis of GFP+ cells and GFP− cells in chimeric E8.5 embryo derived from GFP-labeling p53 KO-1 cells. The percentage of the 1n (G0/G1) peak from p53 KO-1 chimeric E8.5 GFP+ cells was 25.3%.
(E) Expression levels of pluripotency genes (Oct4, Nanog, Stella, Rex1, and Klf4) in chimeric E8.5 haploid cells from GFP+ cells, with WT-haESCs as a loading control (n = 3 independent experiments). Data presented as mean ± SEM. t test: ∗∗p < 0.01, ∗∗∗p < 0.001.
(F) Morphology of chimeric embryos at E8.5 derived from GFP-labeling p53 KO-diploid cells.
(G) Morphology of chimeric embryos at E10.5 derived from GFP-labeling p53 KO-1 cells. WT E10.5 was used as a control.
(H) DNA-content analysis of GFP+ cells and GFP− cells in chimeric E10.5 embryo derived from GFP-labeling p53 KO-1 cells. The percentage of the 1n (G0/G1) peak from p53 KO-1 chimeric E10.5 GFP+ cells was 21.4%.
(I) Teratoma formed from p53 KO haESCs was identified by H&E staining. Scale bar, 100 μm. The tissues shown were gut epithelium (endoderm), muscle (mesoderm), and neural tissue (ectoderm).
(J) DNA-content analysis of teratoma. The percentage of the 1n (G0/G1) peak from all the tissue was 3.1%.
(K) Expression levels of pluripotent genes (Oct4, Nanog, Stella, Rex1, and Klf4) in the 1n cells sorted from teratoma determined by qPCR, with WT-haESCs as a loading control (n = 3 independent experiments). Data presented as mean ± SEM. t test: ∗∗∗p < 0.001.