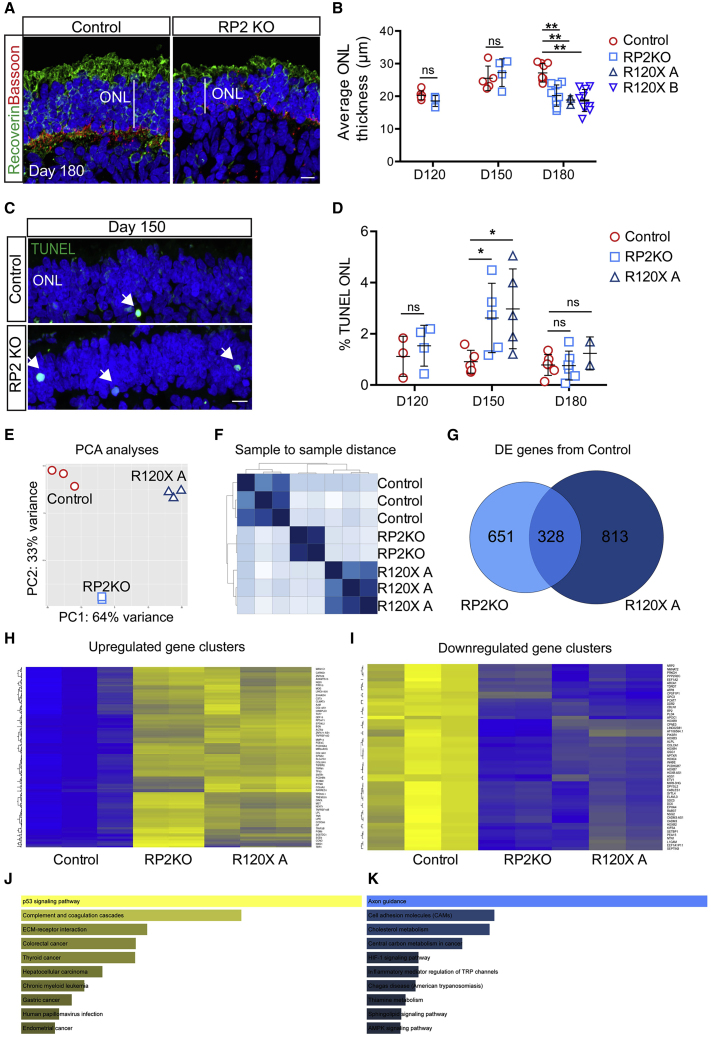
Figure 2.
Photoreceptor Differentiation-Associated Cell Death in RP2 KO Organoids
(A) ICC of control and RP2 KO retinal organoids showing reduced ONL thickness in RP2 KO. Recoverin staining demarcates the ONL terminating in the synaptic layer stained with Bassoon. Scale bar, 10 μm.
(B) Mean ONL thickness per organoid was measured from tilescans of cryosections of a whole organoid at D120, D150 (n = 5 control; n = 4 RP2 KO at both time points), and D180. Significant ONL thinning was recorded at D180 in RP2 KO (n = 9 independent organoids) and R120X lines (n = 3 R120X A organoids, n = 9 R120X B organoids), but not in controls (n = 10 independent organoids; p ≤ 0.01; mean ± SD).
(C) TUNEL reactive nuclei (arrows) in the ONL of RP2 KO and isogenic control organoids at D150. Scale bar, 10 μm.
(D) Quantification of TUNEL reactivity. RP2 KO organoids had a significantly higher proportion of TUNEL-positive cells at D150 (n = 5 independent organoids p ≤ 0.05, mean ± SD) but not at D120 (n = 3 control; n = 4 RP2 KO) or D180 (n = 5 control; n = 6 RP2 KO). R120X organoids also had increased TUNEL reactivity at D150 (n = 5 at D150 n = 2 at D180 independent organoids).
(E) Principal-component analyses of RNA-seq data from ROs (n = 3 control and R120X A, n = 2 RP2 KO independent organoids).
(F) Sample to sample distance between samples.
(G) Venn diagram showing differentially expressed genes between RP2 KO and control and R120X and control and common genes.
(H) Heatmap showing upregulated clusters of differentially expressed genes (blue, lower expression; yellow, higher expression).
(I) Heatmap of downregulated clusters of differentially expressed genes.
(J) KEGG pathway analyses of upregulated genes.
(K) KEGG pathway analyses of downregulated genes.