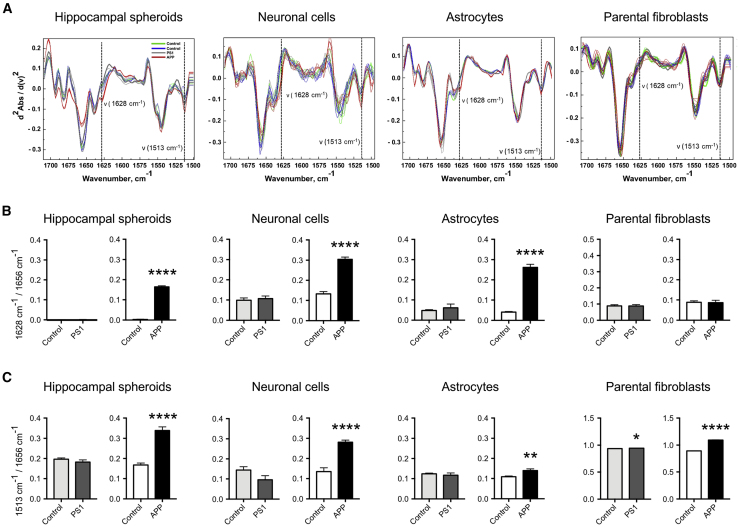
Figure 4.
APP Variant HSs Exhibit Increased Protein Aggregation
(A) Normalized second derivatives of the infrared light absorbance spectra in Amide I and II regions. Dashed lines and arrows indicate the increased protein aggregation as elevation of β sheet structures (1,628 cm−1) and structural changes corresponding to protein aggregation, as elevation at the band positioned at 1,513 cm−1 in APP in HSs at DIV 100, hippocampal neurons at DIV 56, astrocytes at DIV 120, and parental fibroblasts.
(B) Bar diagrams reflecting the β sheet structure content as shown by the absorbance ratios 1,628 to 1,656 cm−1 in the same cells described in (A). Results are presented as mean ± SEM. n = 5–10 spectra per genotype for HSs, n = 20–26 spectra per genotype for parental fibroblasts, n = 3 independent differentiations per genotype for hippocampal neurons and astrocytes. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001. Statistical analysis by two-tailed t test.
(C) Bar diagrams reflecting structural changes as shown by the absorbance ratios 1,513 to 1,656 cm−1 in same cells described in (A). Results are presented as mean ± SEM. n = 5–10 spectra per one genotype for HSs. n = 20–26 spectra per genotype for parental fibroblasts, n = 3 independent differentiations per genotype for hippocampal neurons and astrocytes. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001. Statistical analysis by two-tailed t test.