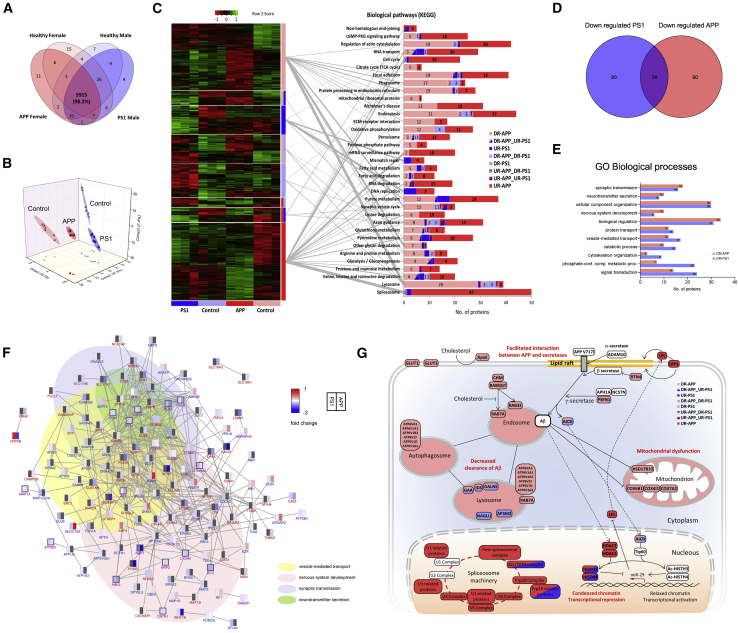
Figure 5.
Label-free Quantitative Proteomics Reveals Important Alterations in the Proteome of APP and PS1 Variant HSs
(A) Venn diagram representing the number of proteins identified in HSs. The numbers correspond to the set of three different processes of generating HSs (biological replicates) and two technical replicates of the LC-MS/MS analysis.
(B) Principal component analysis using the normalized intensities of proteins identified/quantified in all samples.
(C) Heatmap of the protein relative abundances that were significantly dysregulated in at least one of the variant samples (left). n = 3 independent differentiations per genotype, n = 2 LC-MS/MS analyses. False discovery rate (FDR) < 0.05 was considered significant. Statistical analysis by two-tailed t test. Biological pathways (Kyoto Encyclopedia of Genes and Genomes) enriched in at least one of the eight blocks of dysregulated proteins (right). The graph shows the distribution of the proteins involved in each biological pathway by blocks of dysregulated proteins. Gray lines connect each block of proteins with their most enriched pathways.
(D) Venn diagram representing the number of synaptic proteins significantly downregulated in PS1 and APP variants when compared with their gender controls. n = 3 independent differentiations per genotype, n = 2 LC-MS/MS analyses. p < 0.05 with log2 fold change <−0.2 was considered significant. Statistical analysis by two-tailed t test.
(E) Representative enriched biological processes (Panther GO-Slim) for the downregulated synaptic proteins in PS1 and APP variants are shown with the number of corresponding proteins.
(F) Interaction network of the synaptic proteins downregulated in at least one of the samples carrying a mutation (n = 206). Each protein is represented by a rectangle divided into two parts (left-right), where different colors specifying the corresponding fold changes in PS1 and APP variants compared with their gender controls. Lines between the nodes (rectangles) indicate protein-protein interactions. Nodes with dark gray border represent synaptic proteins that are significantly downregulated in each variant. Purple label indicates significant downregulation in both variant, while blue and red labels indicate significant downregulation in PS1 or APP variant, respectively. n = 3 independent differentiations per genotype, n = 2 LC-MS/MS analyses. p < 0.05 with log2 fold change <−0.2 was considered significant. Statistical analysis by two-tailed t test.
(G) Schematic summary of AD-related dysregulated pathways and proteins in APP and PS1 variant HSs revealed by quantitative proteomics.