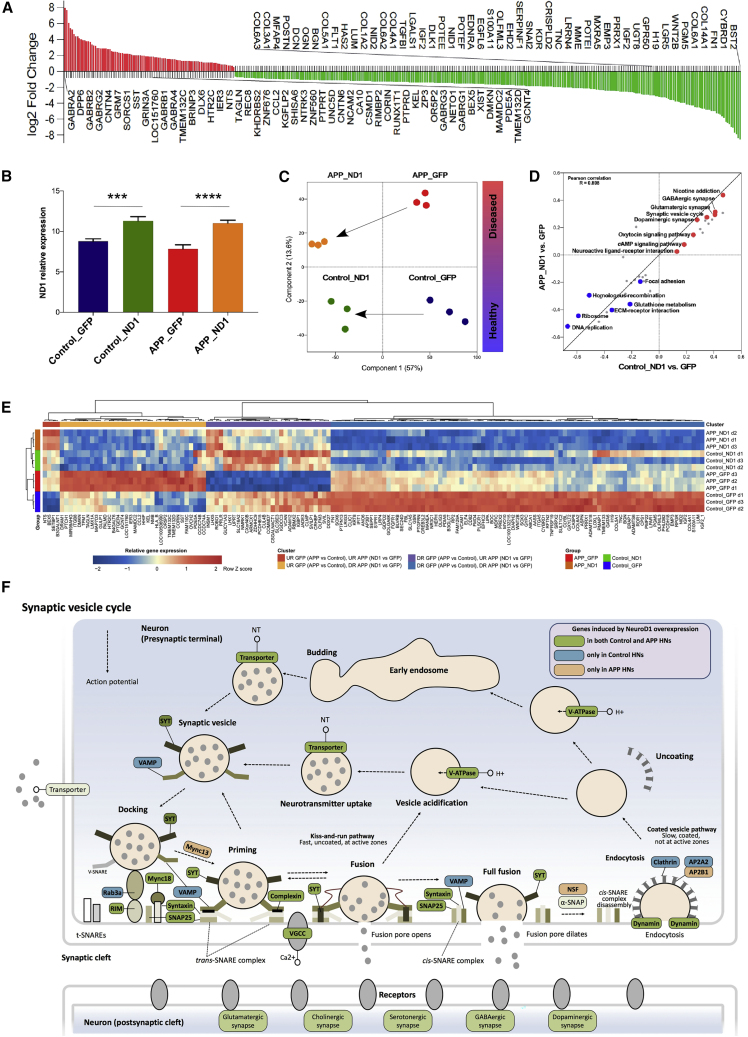
Figure 6.
ND1 Ex Vivo Gene Therapy Modulates Transcription of Genes Relevant to AD
(A) Genes dysregulated in APP variant hippocampal neurons at DIV 50. The top 50 genes upregulated (in red) and downregulated (in green) are annotated. n = 3 independent differentiations per genotype. FDR < 0.05 was considered significant. Statistical analysis by two-tailed t test.
(B) Relative expression levels of ND1 gene. Results are presented as mean ± S.D. n = 3 independent differentiations per genotype. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Statistical analysis by one-way ANOVA followed by Tukey's post hoc test.
(C) Principal component analysis using gene expression profiles of hippocampal neurons.
(D) 2D annotation enrichment analysis for biological pathways based on the differential expression profiles of transcripts between hippocampal neurons transduced with ND1 and GFP, in both control (x axis) and APP variant (y axis). Pathways significantly enriched after ND1 overexpression are highlighted, downregulated (blue) and upregulated (red). n = 3 independent differentiations per genotype. Enriched pathways were selected according to a Benjamini-Hochberg FDR of 0.01 (Table S5).
(E) Heatmap of the relative expression of genes that were significantly dysregulated in APP variant hippocampal neurons and modulated by ND1 overexpression (Table S5). n = 3 independent differentiations per genotype. Statistical analysis by two-tailed t test.
(F) Schematic representation of synaptic vesicle cycle pathway in hippocampal neurons. Genes whose expression was induced by ND1 overexpression are highlighted.