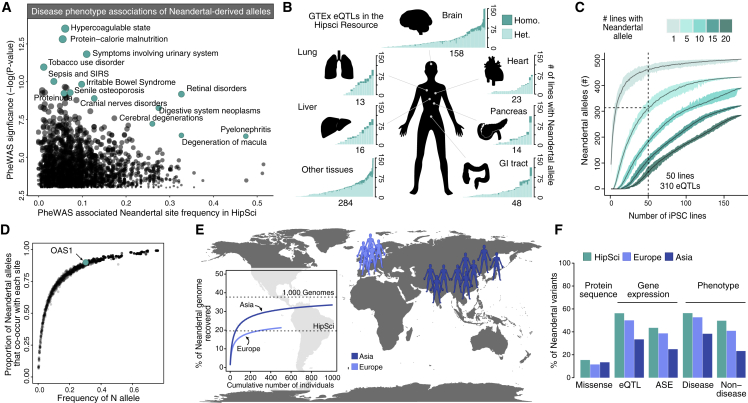
Figure 2.
The HipSci Resource Harbors Extensive Functionally Relevant Neandertal DNA
(A) The frequency of single nucleotide changes introgressed from Neandertals that have significant associations with human disease phenotypes (Simonti et al., 2016), with select associations highlighted in teal.
(B) Barplots illustrating the number of iPSC lines that contain each Neandertal-associated eQTL (y axis sorted by frequency of alleles in HipSci), from the GTEx dataset present in the HipSci resource, colored by tissue and by homozygous (dark) or heterozygous (light). Neandertal-associated eQTLs in the brain represent the pool of Neandertal-associated eQTLs that are found in at least one of the 13 GTEx brain tissues.
(C) A power analysis shows the average of how many Neandertal eQTLs are present in at least a certain number of cell lines (color) out of a random sample of X lines (x axis) from the HipSci resource (grey). The range across the random samplings is shown in respective colors for each number of lines.
(D) Plot showing the co-occurrence of Neandertal alleles within individuals. The frequency of each Neandertal allele is plotted against the proportion of all other Neandertal alleles with which it co-occurs in the HipSci resource. For example, an OAS1 Neandertal-derived allele is found at relatively high frequency (0.33, and present in 53% of all individuals) in the HipSci resource and is therefore paired with the majority of other Neandertal alleles.
(E) Inferred Neandertal haplotypes in the 5 European (light blue, total of 505 individual) and 10 Asian (dark blue, total of 1,009 individuals) populations of the 1000 Genomes Project (1000 Genomes Project Consortium et al., 2015) recovered 21.3% and 33.5% of the Neandertal genome in Europeans and Asians, respectively.
(F) Barplots show the presence of putative functional Neandertal variants at frequencies greater than 5% in the HipSci cohort as well as 1000 Genomes European and Asian populations (allele-specific expression [ASE]). We found that amino acid-modifying variants were slightly more prevalent in Asian populations, consistent with the higher amount of recovered Neandertal DNA in that population. However, other functional variants derived from association studies with gene expression and phenotype data were more often found in Europeans and the mostly European HipSci individuals, consistent with a detection bias inherited from the association analyses that were conducted in cohorts with a mostly European ancestry.
See also Figure S1.