Figure 4.
Tracking RNA from Neandertal-Introgressed Haplotypes during Cortex Development
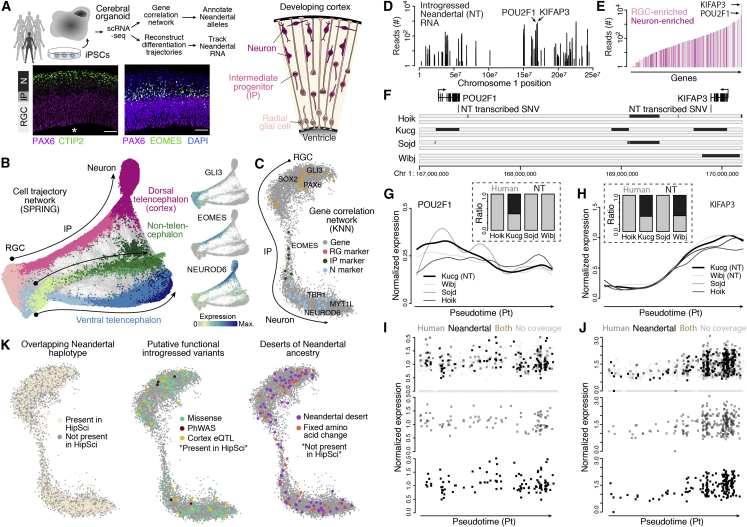
(A) Overview of analysis using single-cell RNA sequencing (scRNA-seq) data from cerebral organoids from multiple individuals with whole-genome genotype data. Schematic and immunohistochemistry show the structure of cortical-like regions within cerebral organoids. Radial glial (RG), intermediate progenitor (IP), and neuronal (N) (Mora-Bermúdez et al., 2016) cells were extracted from the organoid scRNA-seq data, and used to identify correlated gene expression patterns during cortical neuron differentiation. scRNA-seq data is from Kanton et al. (2019), and stainings are from Mora-Bermúdez et al. (2016).
(B) SPRING reconstruction based on organoid scRNA-seq data from seven individuals (including four HipSci iPSC lines), with clusters colored by cell types. All subsequent analysis was based on RGCs, IPs, and neurons in the cortical trajectory.
(C) A correlation network (knn, k = 70) using 7,349 genes (gray) that highly correlate in expression (r > 0.6) with transcription factors that have been shown to regulate progenitor proliferation/self-renewal and neuron differentiation (Camp et al., 2015) is shown with transcription factors that represent individual cell-type-specific expression colored (orange, RG; green, IP; blue, N).
(D) Based on the reads from the scRNA-seq that overlap informative positions we are able to classify the reads as “Neandertal” or “modern human” depending on the observed allele at these sites. Plot shows the distribution of the number of reads with Neandertal informative variants across chromosome 1 from scRNA-seq data from organoids from four HipSci lines. Reads assigned to KIFAP3 and POU2F1, two genes that are part of the gene expression correlation network shown in Figure 3, are highlighted. POU2F1 and KIFAP3 show the highest expression in progenitors and neurons, respectively.
(E) Number of reads covering Neandertal-informative variants which shows differences in their genotype between the 4 HipSci individuals for 535 genes. Bars representing genes with an expression difference of at least 2-fold between progenitors and neurons are highlighted in light pink (larger expression in progenitors) and dark pink (larger expression in neurons). Genes with no expression differences are shown in gray.
(F) Gene models for KIFAP3 and POU2F1 are shown together with the locations of the Neandertal-informative SNPs, rs4519 and rs1059761, in the corresponding 3′ UTRs of these genes. The lower panel shows inferred Neandertal haplotypes in the four HipSci individuals overlapping chr1:167,000,000-170,000,000.
(G and H) Spline-interpolated gene expression trajectories across pseudotime for four HipSci cell lines for POU2F1 (G) and KIFAP3 (H). Cell lines with a chromosome carrying Neandertal haplotypes overlapping KIFAP3 and POU2F1, respectively, are labeled with “NT.” Barplots show the fraction of reads for each cell line carrying at the Neandertal-informative position the modern human (gray) and Neandertal (black) variants.
(I and J) The top panel shows the expression of all kucg cells for POU2F1 (I) and KIFAP3 (J) and whether cells have reads assigned that carry a Neandertal variant (black), a human variant (gray), both Neandertal and human variants (brown), or have no reads carrying Neandertal-informative variants (light grey). The bottom two panels highlight the cells with human-only (gray) or Neandertal-only reads.
(K) Cortical gene correlation network. Left: 1,777 genes (24% of network genes) with overlapping Neandertal haplotypes in the HipSci individuals are colored in beige. Middle: subset of genes with overlapping Neandertal haplotypes in HipSci that carry Neandertal variants that are potentially functional, including eQTLs in cortical brain regions that are linked to a Neandertal haplotype in GTEx (Dannemann et al., 2017), variants that change the amino acid sequence, and variants that have been associated with clinical phenotypes (Simonti et al., 2016). Right: among the genes without overlapping Neandertal haplotypes in the HipSci are 81 genes that are located in desert regions devoid of Neandertal ancestry (Vernot et al., 2016) and 33 genes with fixed human-derived amino acid substitutions (Prüfer et al., 2014).
See also Figures S2–S4.