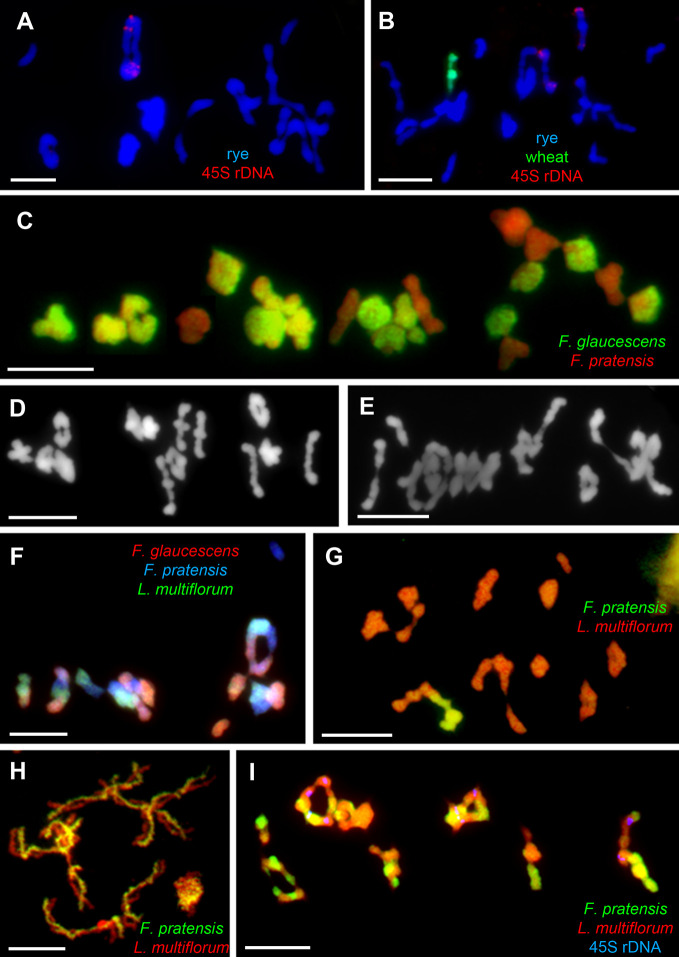
Figure 2.
Chromosome assocaitions in allo- and autopolyploids from the Poaceae family. Chromosome pairing in autotetraploid rye (2n = 4x = 28, RRRR) differs depending on the presence or absence of Ph1 located on the introgressed 5BL chromosome arm of wheat. In (A), trivalents and quadrivalents are commonly observed in the control line (2I+4II+2III+3IV), in (B), multivalent chromosome formation is reduced in the line (6I+7II+2IV), where 5B and 5BL are introgressed. In both (A, B), genomic DNA of Triticum aestivum was labeled with digoxigenin (green coloring), 45S rDNA was labeled with biotin (red), and genomic DNA of Secale cereale served as blocking DNA; all chromosomes counterstained with DAPI (blue). In (C), the chromosome-pairing control system similar to that of Ph1 found in allohexaploid Festuca arundinacea (2n = 6x = 42) hampers the associations of homeologous chromosomes and multivalent formation (21II). Genomic DNA of F. glaucescens was labeled with digoxigenin (green), while genomic DNA of F. pratensis was used as blocking DNA; all chromosomes were counterstained with DAPI (red pseudocolor). In (D), the homoeolog suppressor was probably inherited from one of the progenitors, F. glaucescens, as this species also forms only bivalents during meiosis (14II). Conversely, in (E), multivalent formation was detected in the autotetraploid form of the other progenitor, F. pratensis (2I+7II+3IV). The system is hemizygous-ineffective, thus allowing for promiscuous homeologous chromosome associations in tetraploid hybrids of F. arundinacea × Lolium multiflorum, where only one copy of the gene(s) is present (F). Here, genomic DNA of F. glaucescens was labeled with biotin (red coloring) and that of L. multiflorum labeled with digoxigenin (green), while that of F. pratensis was used as blocking DNA; all chromosomes were counterstained with DAPI (blue). In (G), homeologous chromosomes of F. pratensis and L. multiflorum pair freely in the substitution lines (1I+8II+1III+2IV) as well as in diploid Festuca × Lolium hybrids (7II), as seen in diplotene shown in (H), due to the absence of any chromosome pairing system and the phylogenetic relationship of both genomes. Note many chiasmata between homeologous chromosomes. This results in frequent homeologous recombinations and massive chromosome rearrangements in successive generations (I), as can be seen in the tetraploid L. multiflorum × F. pratensis cv. ‘Sulino’ (7IV). In panels (G–I), genomic DNA of F. pratensis was labeled with digoxigenin (green coloring), while genomic DNA of L. multiflorum served as blocking DNA and all chromosomes were counterstained with DAPI (red pseudocolor).