Figure 7 |.
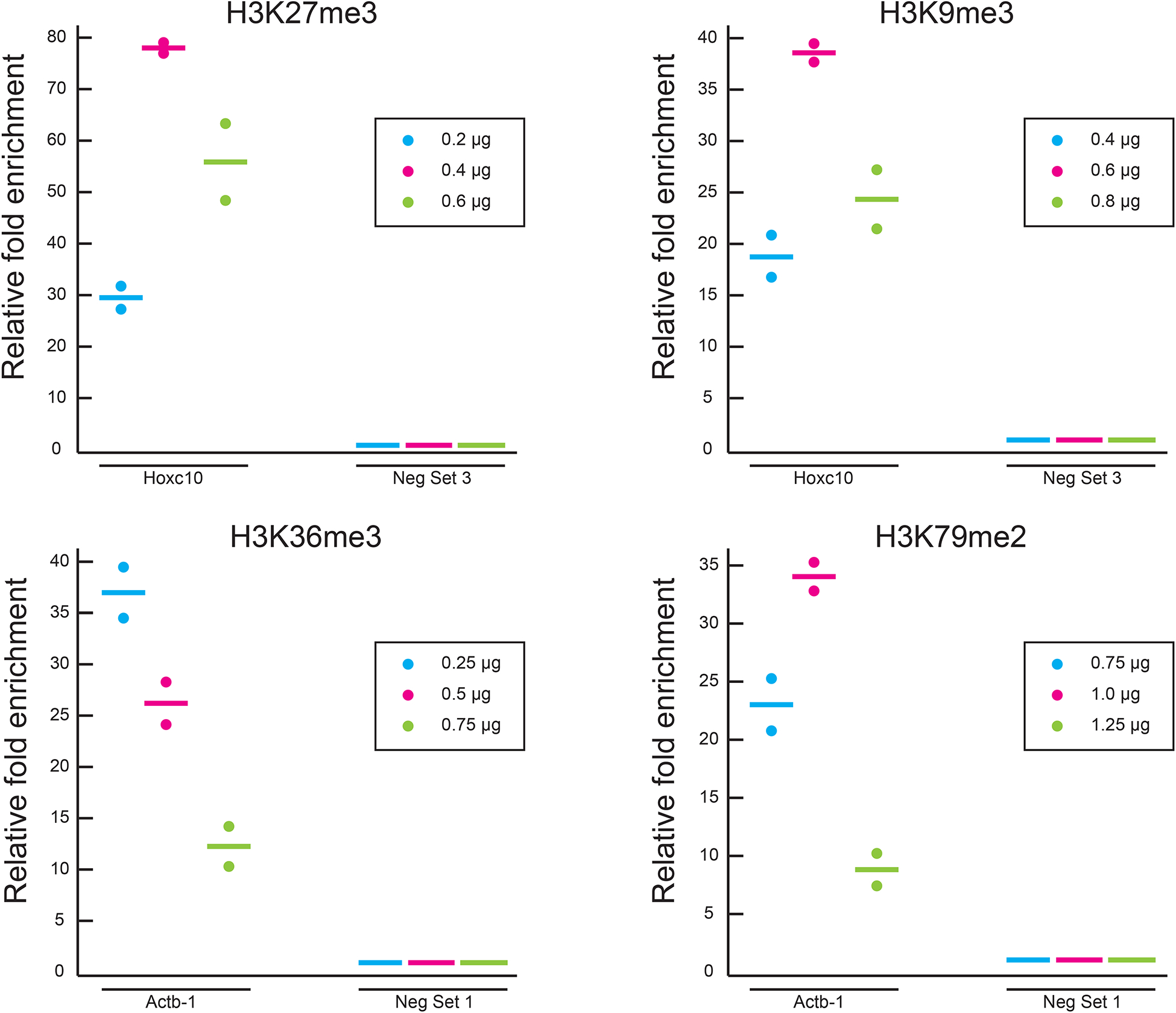
Optimization of antibody quantity using MOWChIP-qPCR for 4 histone marks. For each histone mark, 3 different quantities of antibody were used during IP bead coating and enrichment of ChIP DNA at positive (Hoxc10 for H3K27me3 and H3K9me3; Actb-1 for H3K36me3 and H3K79me2) and negative (Neg Set 3 for H3K27me3 and H3K9me3; Neg Set 1 for H3K36me3 and H3K79me2) loci. Enrichment was examined by qPCR under each condition. All experiments were conducted in duplicate and the horizontal lines represent the mean. The relative fold enrichment was normalized against that of Neg Set 3 or Neg Set 1. 1000 nuclei from mouse prefrontal cortex were used in each assay and MNase digestion was used for chromatin fragmentation. 10-week old CD-1 male mice were used in the study. The nuclei were not sorted and included neurons and glia. The animal protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of Virginia Commonwealth University.
