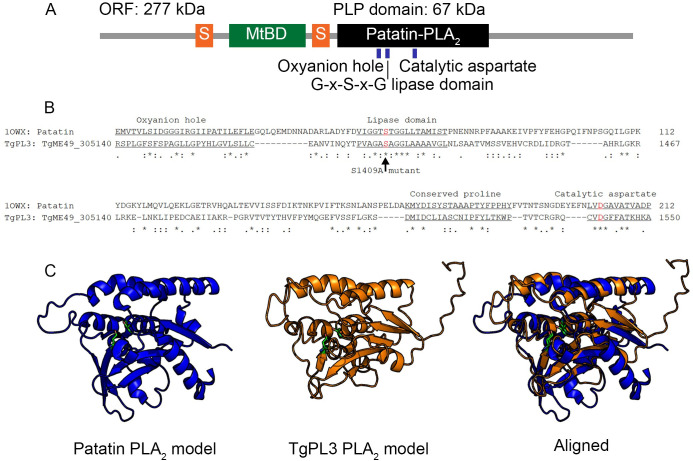
Fig 1. Predicted domains of TgPL3 based on sequence and structural alignments.
(A) In orange, two serine-rich domains (S) flank the microtubule binding domain (MtBD) in green. The first serine-rich domain contains 34 consecutive serines and the second contains 37 serines interrupted by 2 prolines and 2 alanines. The putative PLP domain is shown in black. (B) The PLP domain contains all the necessary conserved amino acids for activity based on sequence alignment. The known conserved patatin motifs are underlined and catalytic S/A dyad are colored red. The serine to alanine mutation (S1409A) is indicated by an arrow. (C) Structural alignment of the TgPL3 PLP domain (orange) to the patatin PLA2 crystal structure (blue) predicts correct folding to form the active site. The secondary and tertiary structures of the TgPL3 PLP domain were predicted using I-TASSER and the resulting model was aligned to the PDB model of patatin (1OWX) in PyMOL. The catalytic serine and aspartate dyad are shown in green in each model.