Figure 1Key Figure. Reduced Complexity Crosses (RCCs) for Complex Trait Analysis.
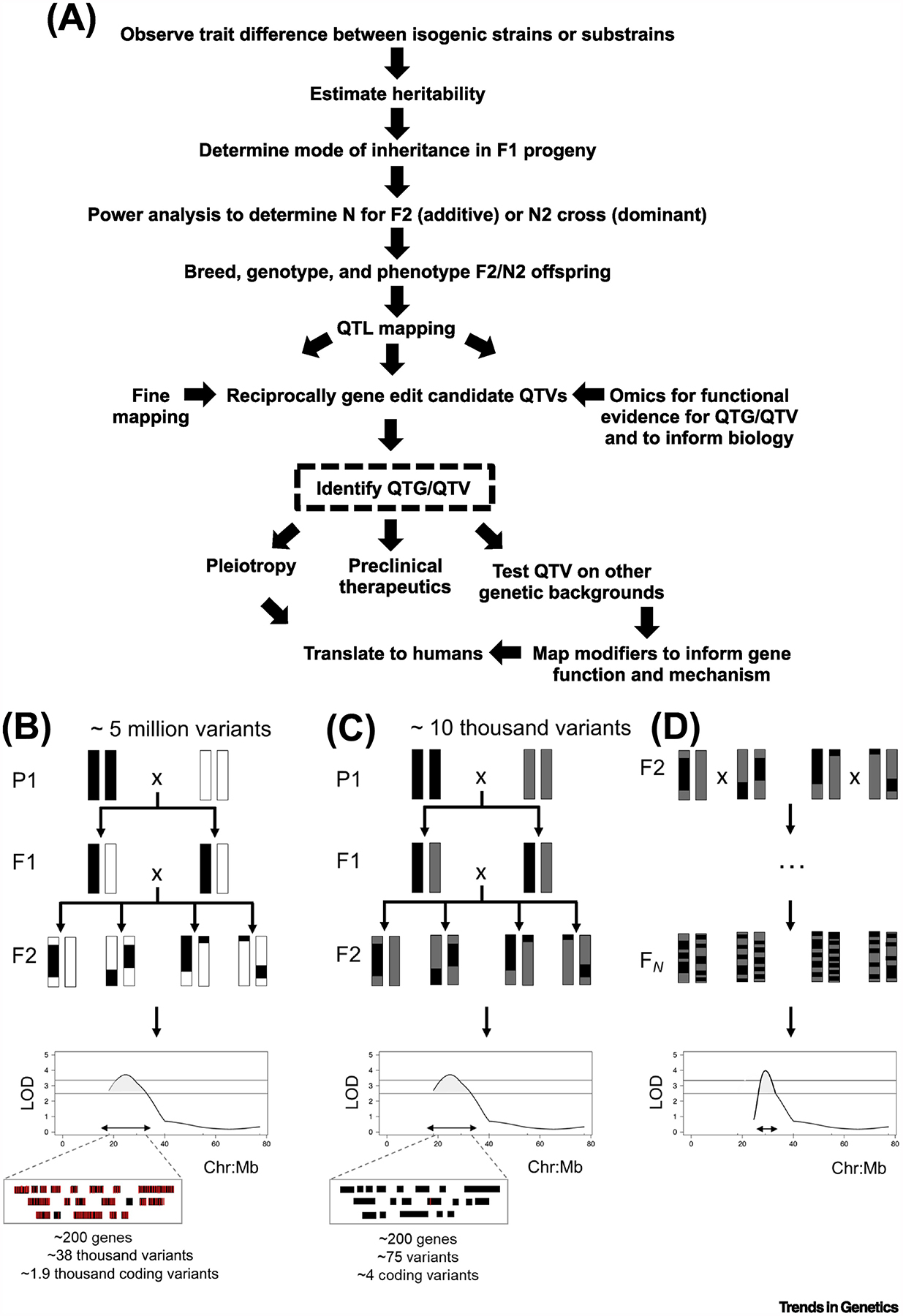
(A) RCC flow chart. (B) A classical F2 intercross between two inbred strains. Isogenic F1 offspring are generated by crossing two inbred strains. Every F1 individual has one chromatid from each parental strain for each chromosome. F1 offspring are intercrossed to generate recombinant F2 individuals. Historically, progenitor inbred strains segregate hundreds of thousands to millions of variants (5 million variants is used as an example here). (C) A reduced complexity F2 intercross. RCCs are generated by intercrossing strains the genomes of which are similar and, thus, segregate orders of magnitude fewer variants compared with more divergent inbred strains. As an example, a RCC between C57BL/6 substrains segregates between 10 000 and 20 000 variants (SNPs plus indels), or in other words, 250 000–500 000-fold fewer variants than a classical F2 cross. Even though the number of historical recombination events and the QTL resolution in an F2 cross are low (~20–40 Mb), the number of candidate causal variants underlying each locus is much smaller. For (B) and (C), the number of genes and variants within each QTL interval was determined based on a genome size of 2.64 B bases, a gene model that included 26 000 genes, and a random distribution model of genes and variants across the genome. (D) A reduced complexity advanced intercross (RCAI). The reduced complexity outcross strain (RCO) addresses the low resolution of an F2 cross by increasing the number of recombination events, which yields a narrower quantitative trait locus (QTL) interval and eliminates the need for fine mapping. The required number of generations for intercrossing to provide sufficient quantitative trait gene (QTG)/quantitative trait variant (QTV) resolution increases as genetic complexity increases.
