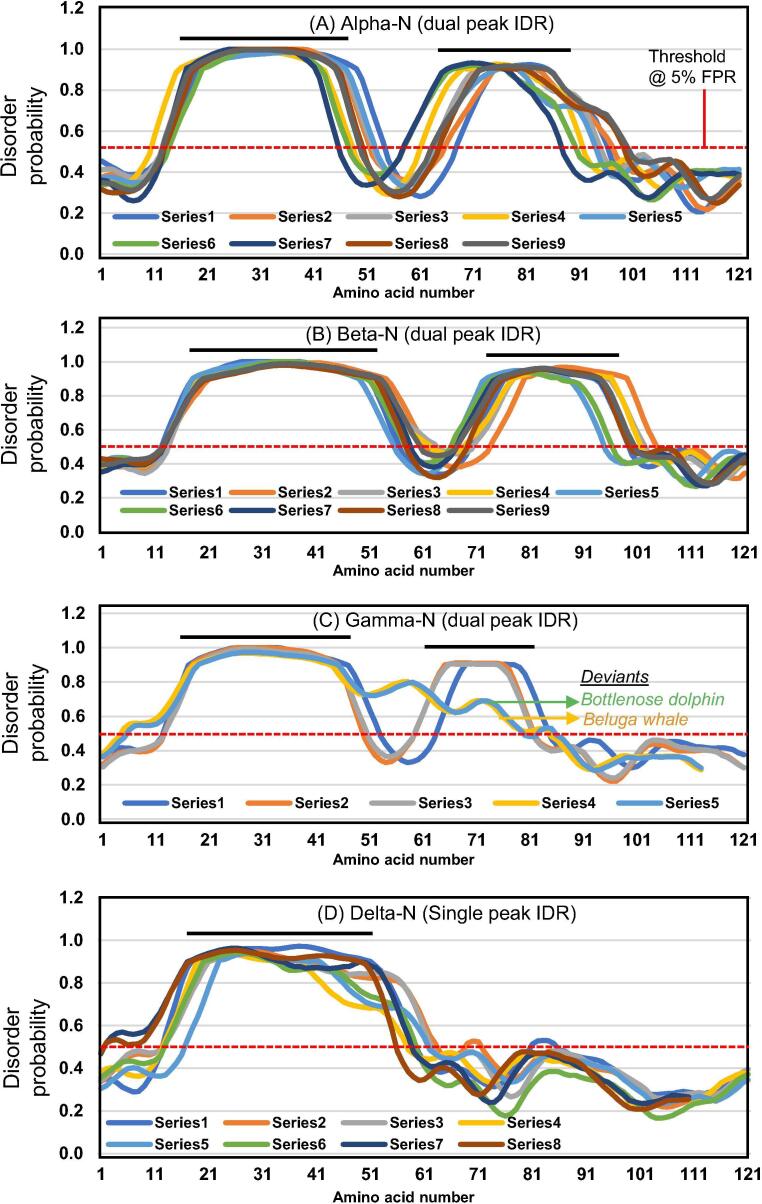
Fig. 1.
Intrinsically disordered region (IDR) at the center of the coronaviral N protein. IDR was predicted by several methods, as described previously [28] and in Materials and Methods, which produced similar results, and those from PrDOS are presented (See Fig. 2 for the corresponding primary structures). The cut-off threshold was set at the relatively stringent FPR (false positive rate) of 5%, corresponding to a disorder probability of 0.5 (Y-axis). Thus, only the areas of disorder probability >0.5 (above the red dotted line) were considered as significant. The “Series” designations of the color-coded graphs are listed in Table 1. As explained in the Materials and Methods, the amino acid numbers (X-axis) do not refer to the full-length protein, but only to the displayed sequence portion, so as to provide a scale of the relative location and length of the two IDR peaks. However, the actual residue numbers are listed in Table 1. Note that alpha, beta, and gamma coronaviruses have two central IDRs, whereas deltacoronaviruses have only one, roughly corresponding to the first. Two highly similar coronaviruses, isolated from beluga whale and bottlenose dolphin, are currently considered in the gamma category [38], [39], but their weak second IDR is marked as ‘deviants’ in this genus. They are presented in the same box as gamma to clearly demonstrate the difference in the peak, but also see Fig. 2, where their sequences are placed in a separate category to indicate this difference. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)