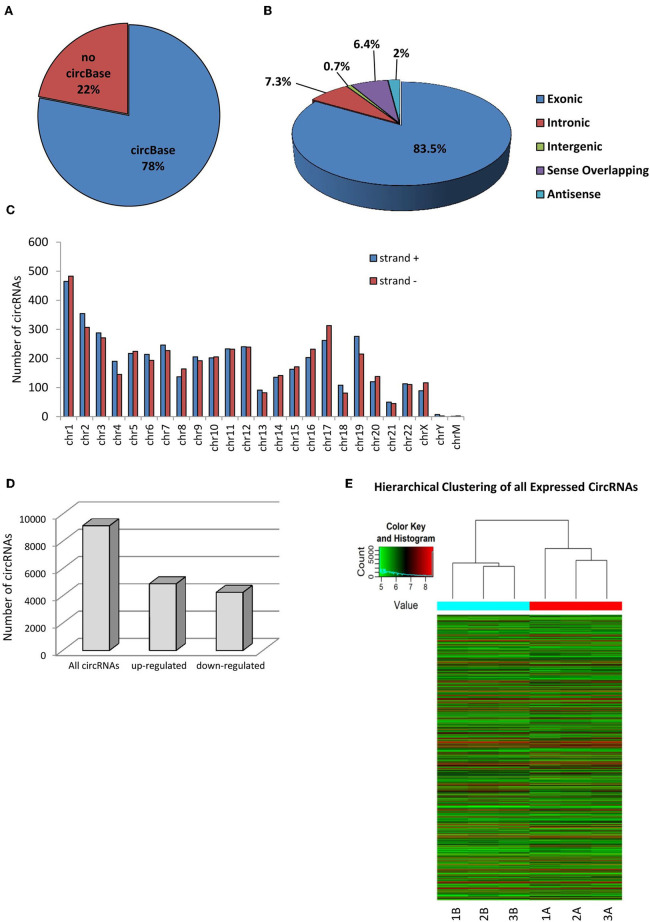
Figure 1.
Overview of circRNA expression in asthenozoospermic derived SPZ. (A) The proportion of circRNAs from circBASE (http://circbase.mdc-berlin.de) and other databases/literature on a total of 9,138 circRNAs identified. (B) The proportion of different types of circRNAs among all predicted circRNAs. (C) Chromosomal distribution of SPZ derived circRNAs, on strand + and strand –. (D) The distribution of up- and down-regulated circRNAs in fraction B compared to fraction A asthenozoospermic derived SPZ among all circRNAs. (E) Hierarchical clustering analysis of total circRNAs samples arranged into two groups, based on their expression levels; in detail, this analysis used different colors to represent the expression values of circRNAs detected in fraction A (n = 3, indicated as 1A, 2A, 3A) and B (n = 3, indicated as 1B, 21B, 3B) SPZ of asthenozoospermic patients.