Figure 1.
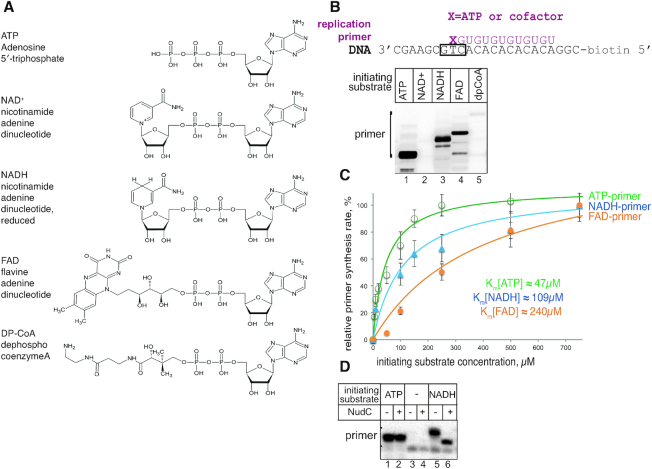
Primase DnaG initiates replication primer synthesis with NAD+, NADH, FAD and DP-CoA. (A) Structures of cofactor molecules (NAD+, NADH, FAD and DP-CoA) in comparison to ATP, the preferred initiating substrate of primase. (B) Replication primer synthesis on single-stranded DNA template, scheme above, with ATP, NAD+, NADH, FAD and DP-CoA as initiation substrates. (C) Plots of the dependencies of the primer amount from concentrations of the initiating ATP, NADH and FAD. Solid lines represent the graphical fits of data (using SigmaPlot software) to the Michaelis–Menten equation. Error bars represent standard deviations from triplicate experiments. (D) RNA primer with 5′-NADH is susceptible to cleavage by NudC hydrolase after DnaG is removed from the complex with high salt wash. Note that absence of ATP from the reaction results in an increased amount of non-specific product, which is not susceptible to NudC in lanes 5, 6. We assume that this band results from initiation with GTP present in the reaction.