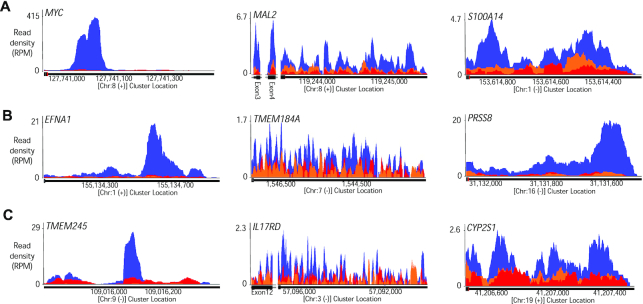
Figure 5.
AGO2 binding RNAseq read clusters for genes that are down regulated, do not significantly change, or are up-regulated. Examples of AGO2 binding clusters identified by AGO2 eCLIP-seq. Blue: Wild type cells. Red: AGO2 knockout cells. Orange: Input control. All clusters are within 3′-UTR regions unless labeled separately (MAL2, IL17RD). Representative AGO2-binding clusters for (A) down-regulated (MYC, MAL2, S100A14) (B) unchanged (EFNA1, TMEM184A, PRSS8) and (C) up-regulated genes (TMEM245, IL17RD, CYP2S1). For inclusion in analysis we required peaks to possess a P value <0.05 and a >4-fold enrichment in read number for wild-type verse AGO2 knockouts.