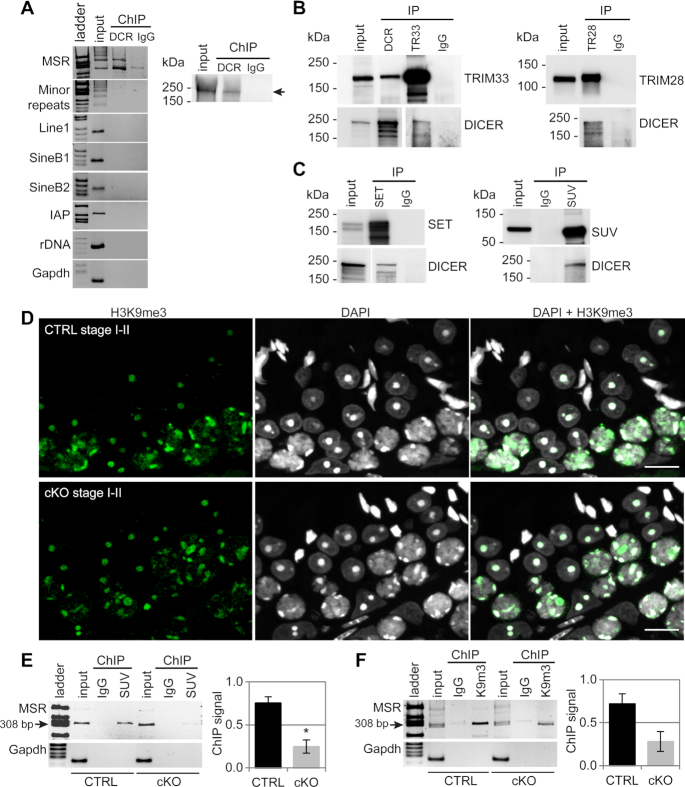
Figure 6.
Epigenetic status of the MSR chromatin is disrupted in Dicer1 knockout testes. (A) Chromatin immunoprecipitation (ChIP) with an anti-DICER (DCR) antibody followed by PCR using primers specific for different repeat sequences revealed the association of DICER with the MSR-containing genomic regions. DICER did not associate with minor satellite repeats, various transposon sequences (Line1, SineB1, SineB2, IAP) or ribosomal DNA repeats (rDNA). Rabbit IgG (IgG) was used as a negative control for immunoprecipitation. The primers for Gapdh gene were used as a negative control for PCR. The top right panel shows immunoblotting with an anti-DICER antibody to validate the performance of the antibody in the ChIP assay. (B) Immunoprecipitation using antibodies against DICER, TRIM33 and TRIM28 followed by western blotting with the same antibodies validated the interaction of DICER (DCR) with TRIM33 (TR33) and TRIM28 (TR28). LYS, lysate; IP, immunoprecipitation; IgG, control IP with rabbit IgG. (C) DICER interacts with histone methyltransferases in the testis. Immunoprecipitation using antibodies against SETDB1 (SET) and SUV39H2 (SUV) followed by immunoblotting with SETDB1, SUV39H2 and DICER antibodies. LYS, lysate; IP, immunoprecipitation; IgG, control IP with rabbit IgG. (D) Localization of H3K9me3 appears unaffected in Dicer1 cKO germ cells. PFA-fixed paraffin-embedded controls (CTRL) and Dicer1 cKO testis sections were immunostained with an anti-H3K9me3 antibody. Scale bar: 10 μm. Panels A-C show a representative figure of an experiment that was independently repeated at least two times. (E) ChIP with anti-SUV39H2 revealed that the association of SUV39H2 with MSRs is reduced in Dicer1 cKO adult testis. (F) Overall H3K9me3 (K9m3) signal on MSR genomic regions appeared reduced in Dicer1 cKO adult testis. The graphs in panels E and F show the quantification of the ChIP-PCR signal (308 bp band) in two independent experiments. The ChIP signal was normalized to the input signal. Signal intensities are shown relative to the CTRL ChIP samples that were set as ‘1’; n = 2, SEM, *P< 0.05. See also Supplementary Figure S5 and Supplementary Table S1.