Figure 1.
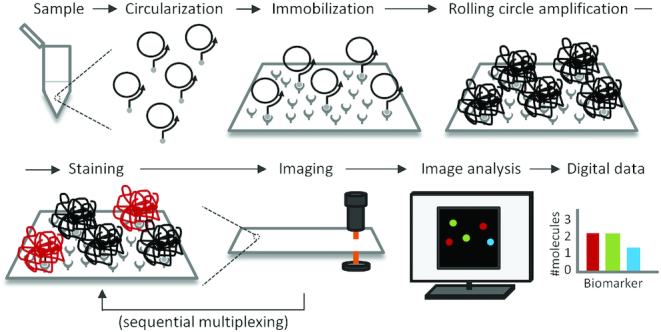
Workflow for digital detection of RCPs. First, a molecular assay results in the formation of DNA circles in response to the presence of target molecules (Circularization) and the DNA circles formed are captured, hybridized to biotinylated primers on a streptavidin coated microscope slide (Immobilization). RCA is then used to elongate the immobilized primers with the DNA circles as templates, resulting in long concatemeric DNA molecules that collapse into individual DNA bundles (rolling circle amplification – RCA). The next step (which can be performed simultaneously with the previous one) is to stain the RCPs with detection oligonucleotides conjugated to fluorophores (Staining), followed by recording the numbers of bright RCPs via fluorescence microscopy (Imaging). Finally, image analysis software is used to digitally enumerate the numbers of RCPs in each image and fluorescence channel (image analysis), followed by processing of the retrieved data using statistics software (data analysis). The staining and imaging steps can be repeated several times for sequential multiplexing. In this case, toehold-mediated DNA strand displacement was deployed to simultaneously de-stain previously imaged RCPs while staining new RCPs to be imaged.
