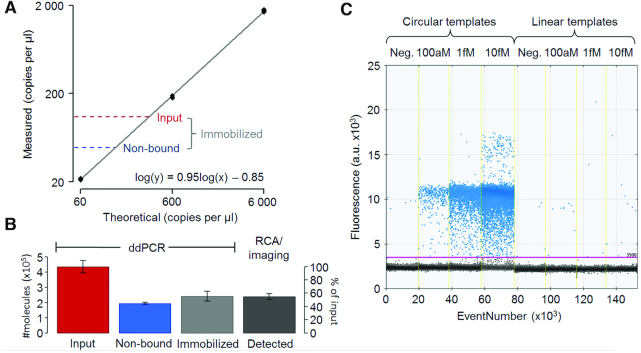
Figure 3.
dPCR was used to evaluate efficiencies for capturing circular DNA molecules on a solid support and for locally amplifying them via RCA. (A) A 10-fold dilution series, analyzed in triplicate technical replicates, demonstrated excellent linearity (black line represents log-log linear regression). The numbers of circular templates added to the solid support (input) and the ones remaining in solution after immobilization (non-bound) were both within the linear range (red and blue dashed lines). (B) The difference between added molecules and ones remaining in solution was used as a measure of the numbers of circular templates immobilized on the solid support (slightly more than 50%). Enumeration via RCA and imaging demonstrated that all immobilized circular templates (light grey bar, defined by dPCR) gave rise to RCPs that could be recorded, as evidenced by the similar heights of the light and dark grey bars. (C) dPCR primers were designed to produce an amplicon over the ligation site so that any remaining uncircularized oligonucleotides would not be amplified. The dPCR specificity for circular templates was verified by analyzing a dilution series of ligated and non-ligated circular templates. Only circularized molecules generated efficient PCR amplification and hence positive droplets.