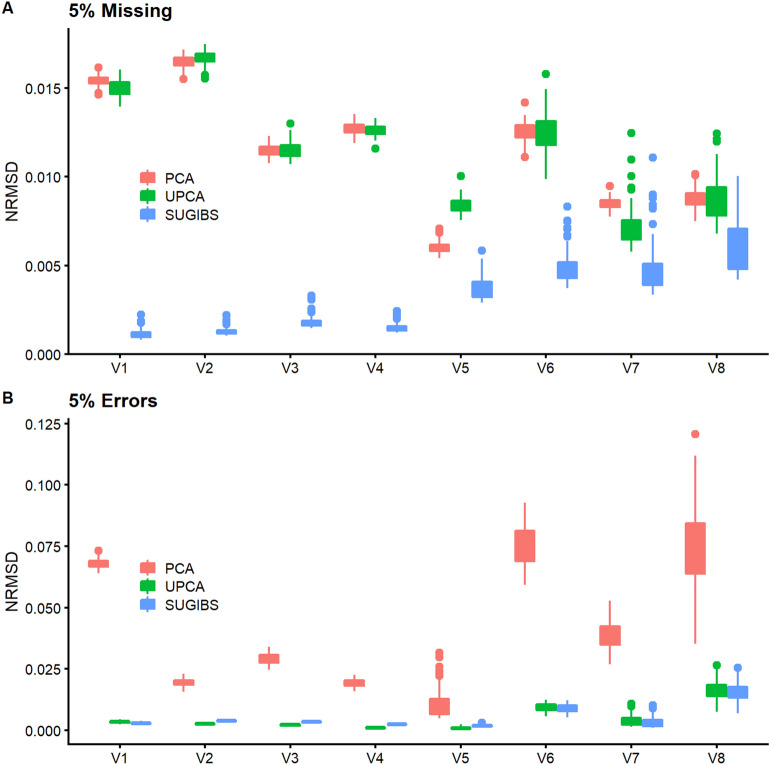
Figure 3.
Normalized root-mean-square deviation (NRMSD) of the top eight axes of PCA, UPCA and SUGIBS. NRMSD measures the root-mean-square differences (RMSD), for the modified HGDP dataset only between the scores on ancestry axes generated using the original genotypes (error free) and the modified genotypes (with simulated errors, A) missing genotypes and (B) erroneous genotypes). The RMSD values were normalized by the range of the ancestry axes generated using the original genotypes, so that NRMSD of the three methods (PCA, UPCA and SUGIBS) are comparable.