Figure 2. RNA sensing and regulatory roles of NLRs.
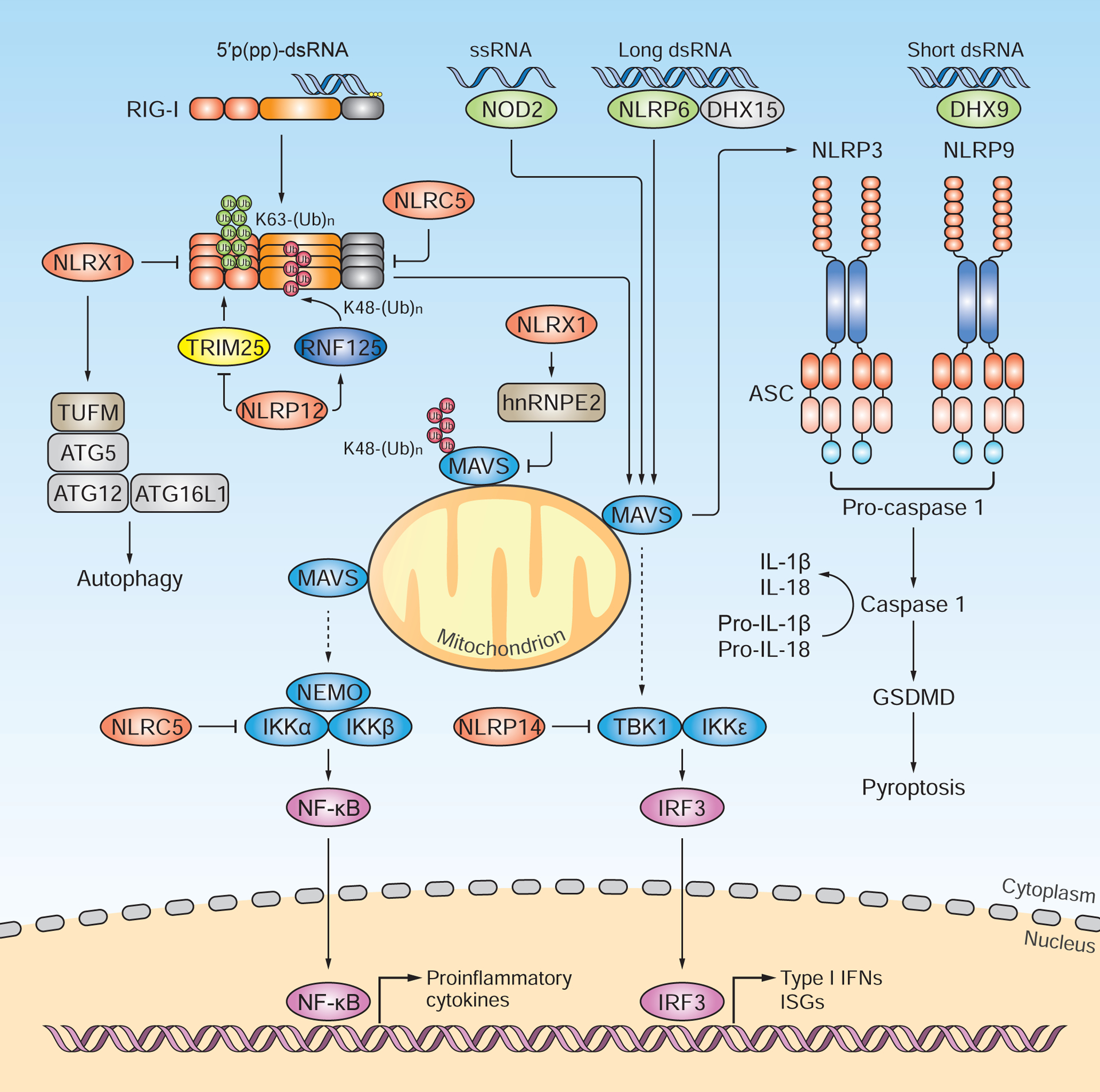
NLRs have been shown to sense viral RNA and trigger type I IFN-and/or inflammasome-dependent host responses, or they regulate other innate sensing pathways such as the RLR pathway. Of note, NLR proteins are expressed and/or function in a highly cell-type-specific or tissue-specific manner, which is not illustrated for simplicity. NOD2 recognizes the ssRNA genome of RSV and mounts MAVS-dependent antiviral responses in hematopoietic and non-hematopoietic cells. NLRP6, in complex with the DEAH-box RNA helicase DHX15, binds viral dsRNA from EMCV and induces MAVS-dependent type I IFN responses in intestinal epithelial cells. Another intestinally-expressed NLR member, NLRP9b, when complexed with DHX9, senses short dsRNA from rotavirus and triggers ASC-and caspase 1-mediated IL-18 production. Additionally, the NLRP9b-DHX9 complex activates caspase-1-dependent pyroptosis through gasdermin D (GSDMD). NLRP12 attenuates RIG-I signaling by counteracting the TRIM25-mediated K63-linked ubiquitination of RIG-I. Moreover, NLRP12 promotes RNF125-mediated K48-linked ubiquitination of RIG-I, which leads to its proteasomal degradation. NLRX1 prevents the RLR-MAVS interaction and also recruits hnRNPE2 to induce MAVS degradation. Furthermore, NLRX1 interacts with TUFM, which promotes autophagy via ATG5, ATG12 and ATG16L1, thereby dampening RLR signaling. NLRP3 interacts with MAVS, and the mitochondrial positioning of NLRP3 promotes inflammasome activation and IL-1β production. NLRC5 and NLRP14 negatively regulate RLR signaling by targeting IKK-mediated and TBK1-mediated innate immune responses, respectively. NLRC5 also binds directly to RLRs and thereby blocks IRF3-dependent cytokine induction. NLRs that promote cytokine responses are shown in green; those that dampen cytokine induction are indicated in red.
