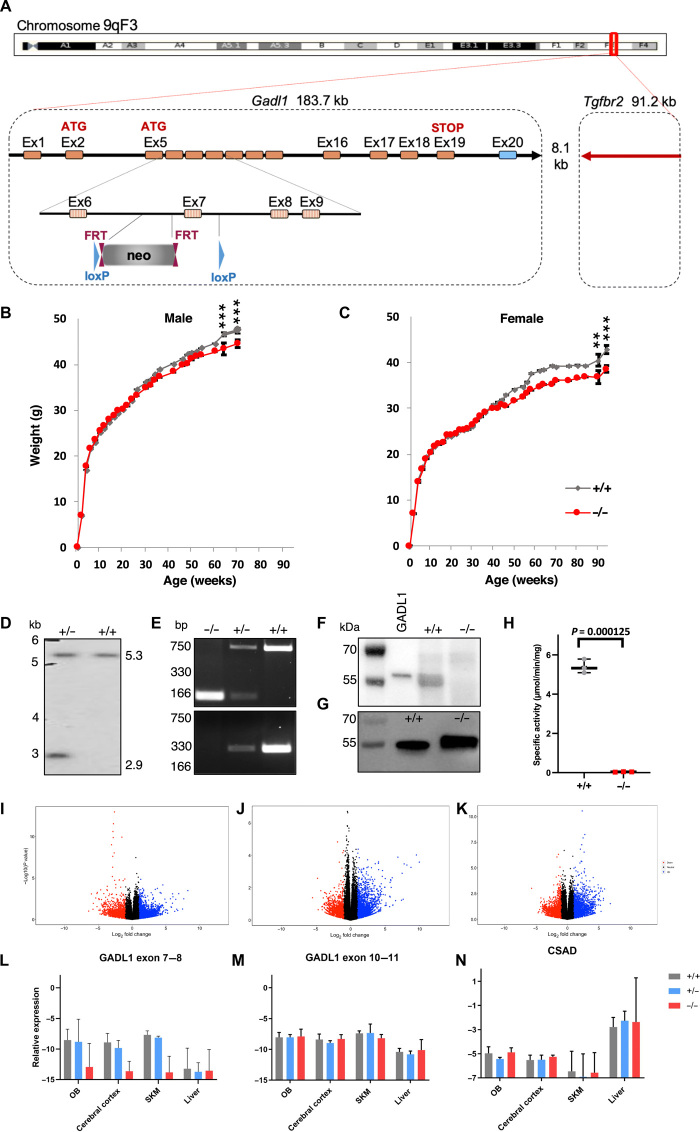
Fig. 1. Generation and characterization of GADL1 KO mice.
(A) Targeting strategy for knocking out exon (Ex) 7 of the mouse Gadl1 locus on chromosome 9. Gadl1 coding sequences (hatched rectangles), noncoding exon portions (blue rectangle), and chromosome sequences (orange rectangles) are represented. The neomycin (neo)–positive selection cassette is indicated between loxP sites (blue triangles) and Flippase recognition target (FRT) sites (plum triangles). (B and C) Growth curves of Gadl1+/+ (n = 4 to 34) and Gadl1−/− mice (n = 4 to 40), presented as means ± SD. Differences between genotypes were significant; P = 0.0008 (64 weeks) and P = 0.0005 (70 weeks) for males and P = 0.0084 (90 weeks) and P = 0.0001 (94 weeks) for females, respectively. (D) Southern blot analysis of genomic DNA from Gadl1+/+ and Gadl1−/−. (E) Genotyping of the offspring from intercrosses of Gadl1+/− mice by polymerase chain reaction (PCR). The DNA band at 166 base pair (bp) is the KO allele (primer 3), while bands at 330 bp (primer 2 and 3) and 750 bp (primer 1) are the wild-type alleles. (F) Representative Western blots of OB samples from Gadl1+/+ and Gadl1−/− mice (34 weeks old, female) using anti-GADL1 antibody. Positive control was recombinant His-tagged GADL1 (2 ng; lane 1). (G) Western blot of recombinant His-tagged truncated Gadl1+/+ and Gadl1−/−. (H) Enzyme activity toward CSA of recombinant His-tagged truncated Gadl1+/+ and Gadl1−/−; P < 0.001. (I to K). RNA expression levels (volcano plots) in OB tissue. (I) Gadl1+/+-to-Gadl1+/− ratio, (J) Gadl1+/+-to-Gadl1−/− ratio, and (K) Gadl1−/−-to-Gadl1+/−ratio. (L to N) Quantitative reverse transcription PCR (qRT-PCR) analysis of normalized mRNA expression in OB, brain, SKM, and liver tissues from 35-week-old female Gadl1+/+ (gray), Gadl1+/− (blue), and Gadl1−/− (red) mice for (L) Gadl1 exons 7 and 8, (M) Gadl1 exons 10 and 11, and (N) CSAD. n = 3 for each genotype. Presented on an Ln y scale as mean of 2ΔCt and upper limit (95%).