Fig. 3.
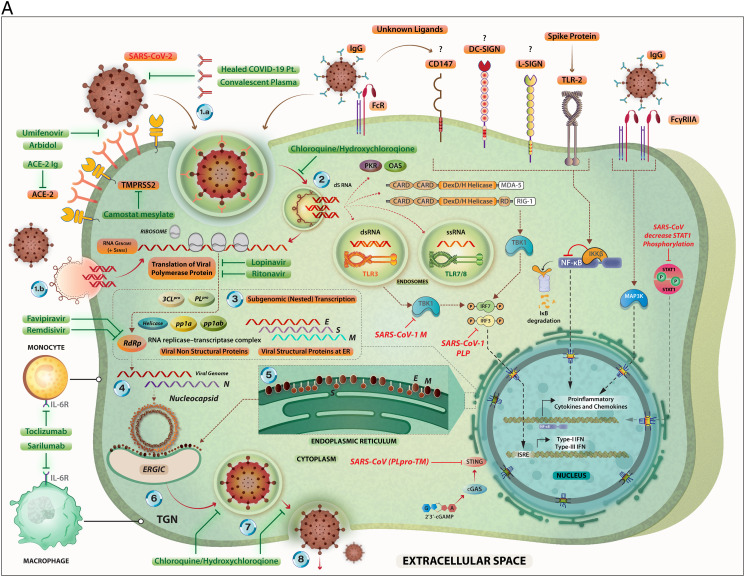
A. The steps in the life-cycle of SARS-CoV-2 are summarized along with the potential mechanism of entry into the host cells. Possible interventions using drugs and their targets during the SARS-CoV-2 life cycle are depicted. Specific signaling proteins that are targets of inhibition to suppress the host immunity are also depicted.
The SARS-CoV-2 can infect its target cells through ACE2-dependent and ACE2-independent (including virus binding to cell surface molecules CD147, DC-SIGN, and L-SIGN; endocytosis of virions or virus-containing apoptotic bodies, and attachment of the virus-coated IgG to FcR). The SARS-CoV-2-related RNA molecules are recognized by intra-cellular PRRs including endosomal TLR3, TLR7 and TLR8; and cytoplasmic sensors including MDA5 and RIG-I. These PRR-mediated signaling pathways eventually may lead to the expression of Type I- and Type III interferons. However, coronaviruses interfere with IFN production through inactivating of the IRF-3. The binding of the SARS-CoV-2-related S protein to the surface TLR2 and the attachment of the virus-coated IgG to FcγRIIA lead to the expression of the pro-inflammatory cytokines and chemokines via induction of the NF-κB and MAPK-related pathways, respectively. The RNA transcription, translation, viral protein synthesis, viral assembling, viral budding to ER, viral transportation into Golgi vesicles, and exocytosis of infective virions are key steps in the cycle life of coronaviruses, that may be targeted by therapeutic agents.
Key to the life cycle of SARS-CoV-2 inside the host cells (monocytes and macrophages along with other cell types): 1.a. Virus entry via ACE-2 mediated endocytosis; 1.b. virus entry through membrane fusion (following binding with ACE-2 and TMPRSS2); 2. release of the viral genome; 3. translation of viral polymerase protein; 4. RNA replication; 5. translation of viral structural proteins (S, M, E) via ER bound ribosomes and nucleocapsid (N) in the cytoplasm; 6. virion assembly at ERGIC; 7. formation of mature virion inside Golgi vesicle; 8. release of infective virions via exocytosis.
The drugs (experimental/repurposed) that are currently prescribed the management of COVID-19 are mentioned above in green boxes. Each of these drugs/antibodies have specific intervention points where they either inhibit- the viral proteins or crucial process like viral entry, translation of viral proteins, assembly of new virions and viral budding, etc. thereby suppressing the multiplication of SARS-CoV-2. An extended list of such drugs is presented in Table 1.
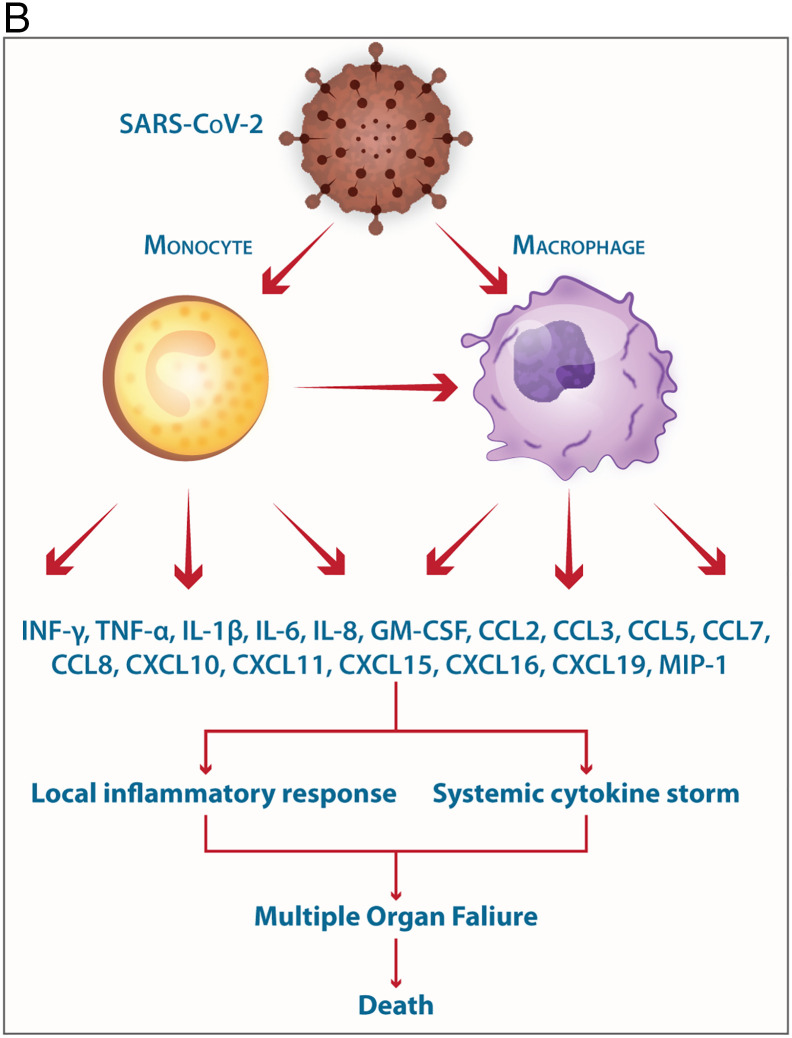
B. Involvement of monocytes/macrophages in the pathogenesis of SARS-CoV-2 the elicitation of mediators of the Cytokine storm is shown.
The mild COVID-19 and moderate COVID-19 were associated with the effective expression of type I IFNs and ISGs in the lungs. Thus, an appropriate local IFN response in the respiratory system can control SARS-CoV-2 infection accompanied by mild and moderate forms of the disease. However, a lower proportion of the SARS-CoV-2-infected patients exhibit severe symptoms. It was proposed, when the viral load is high and the primary local IFN response is failed, the SARS-CoV-2 enters the blood from the lungs and attacks organs expressing high levels of ACE2. The SARS-CoV-2-infected monocytes and macrophages can produce large amounts of numerous types of pro-inflammatory cytokines and chemokines which contribute to the local tissue inflammation and cytokine storm. Both local tissue inflammation and cytokine storm play a key role in the development of COVID-19-related multi-organ failure which causes death in some COVID-19 patients.