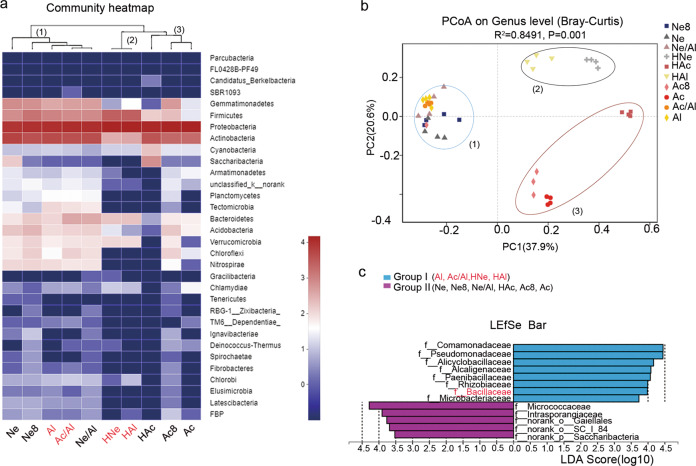
Fig. 4. The rhizosphere microbial community structure was associated with the nodulation of Bradyrhizobium and Sinorhizobium.
a Composition and clustering of bacterial microorganisms from different rhizosphere samples on the phylum level. b PCoA analysis based on Bray–Curtis distances of the rhizosphere microbiomes of soybean seedlings grown in soils shown in Fig. 3a; n = 40. Clustering significance was determined by Adonis (Pr(>F) = 0.001). Acidic soil (Ac), neutral soil (Ne), alkaline soil(Al), acidic soil plus lime (Ac8) and neutral soil plus lime (Ne8), alkaline soil amended with 50% (w/w) acidic soil (Ac/Al) or neutral soil (Ne/Al) and heated acidic soil (HAc), neutral soil (HNe), alkaline soil (HAl). c Linear discriminant analysis (LDA) coupled with the effect size measurements identifies the significant abundance of data in b. Taxa enriched in Group I (purple) and Group II (blue) are indicated with LDA scores, respectively. Only taxa with LDA values greater than 3.5 (P < 0.05) are shown.