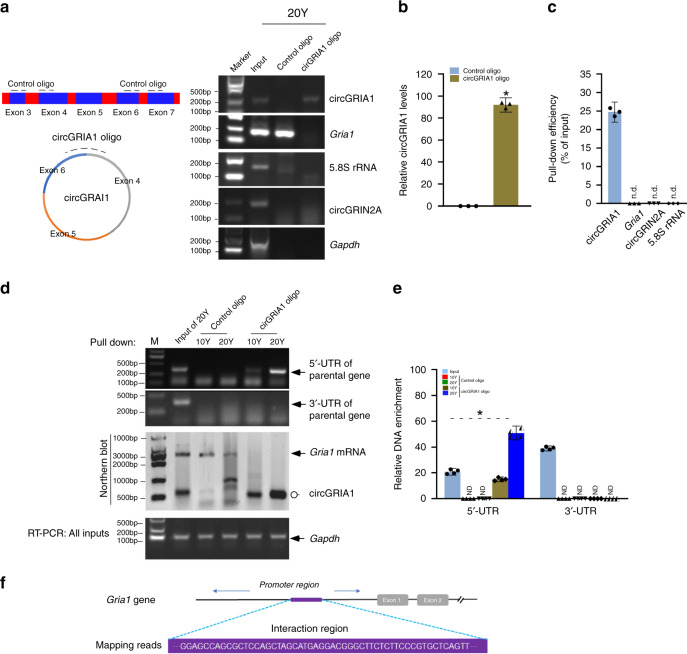
Fig. 3. The regulatory effect of circGRIA1 on the parental genomic locus.
a Biotin-labeled oligonucleotides (oligos) complementary to the junction sequences of circGRIA1 were used for Chromatin Isolation by RNA purification (CHIRP). Oligos complementary to upstream and downstream exons were used as control. Total RNA was extracted from postmortem frozen hippocampal tissues of 20-Y male macaque. The images are representative of three independent experiments of each sample from 2 to 3 animals per age group. b, c Quantification of three repetitions of independent experiment in (a) showed the CHIRP efficiency of circGRIA1. Pre-mRNA of Gria1 was not detected (n. d.) in the CHIRP products as examined with primer pairs corresponding to the parental gene. Both circGRIN2A and 5.8S rRNA were used as controls. Black dash curves represent circRNA junction-targeting (curve) and -nontargeting (linear) sequences of biotin-labeled oligos. d CircGRIA1 interacts with the promoter of its parental gene. Total RNA was extracted from postmortem frozen hippocampal tissues of 10- and 20-year-old male macaques. Upper panel shows PCR products of the parental gene 5′-UTR and 3′-UTR regions against circGRIA1 oligos in ChIRP. Middle panel shows northern blot of Gria1 mRNA and circGRIA1 in the product of CHIRP with digoxin-labeled probes. Semi-quantitative RT-PCR of Gapdh was a loading control. e Quantification of three repetitions of independent experiment in (d). Data represents mean ± S.D.; *p < 0.01, unpaired t-test. f The alignment of circGRIA1-associating motif and the promoter region of its host mRNA gene. A schematic representation of the promoter structure is shown at the top. The gene is coded on the negative strand. The arrows indicate the sanger sequencing data of semi-quantitative RT-PCR products of circGRIA1 parental gene 5′-UTR from CHIRP consistent with the coding sequences. Source data are provided as a Source Data file.