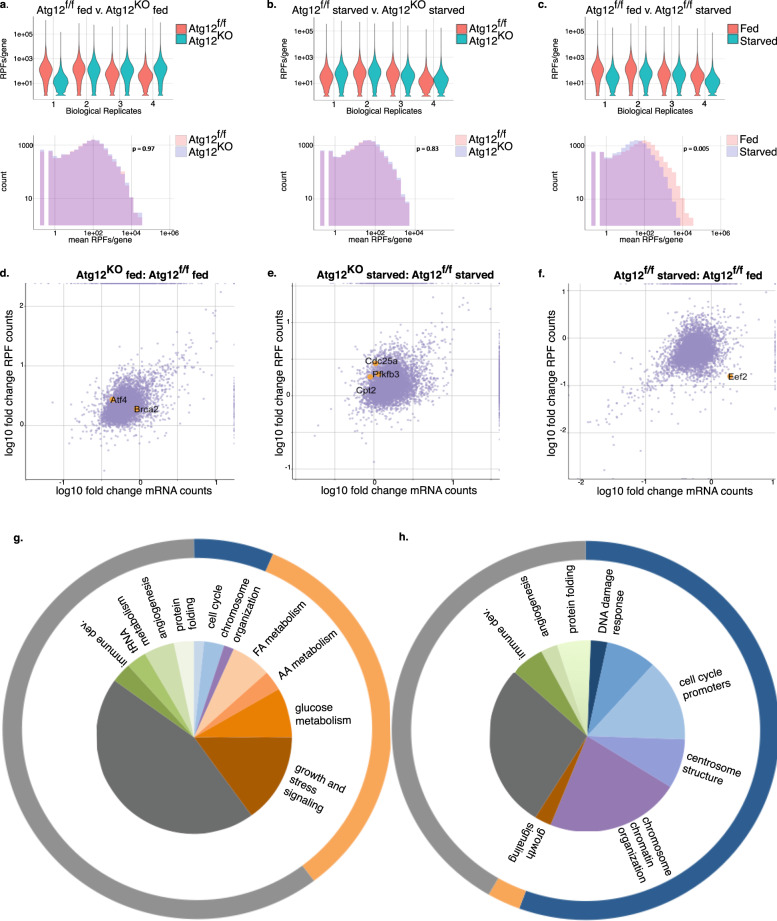
Fig. 3. Ribosome profiling reveals that autophagy supports the translation of proteins involved in DNA damage response and cell-cycle control.
a–c Violin plots of number of read counts of ribosome protected footprints (RPFs) per gene per biological replicate (above) and histogram of the mean of the number of read counts of ribosome protected footprints per gene (below) in (a) Atg12f/f and Atg12KO MEFs in control media (b) Atg12f/f and Atg12KO MEFs following 2 h HBSS starvation or (c) Atg12f/f MEFs in control media or following 2 h HBSS starvation, p = 0.005 by t test. d–f Fold change of RPF counts versus fold change of mRNA counts. Labeled points in orange are mRNAs whose change in ribosome occupancy was significant, and protein level changes confirmed by immunoblotting (see Supplementary Fig. 3c). g, h Molecular functions of mRNAs whose ribosome occupancy is g increased (p < 0.01, n = 36 significant genes) or h decreased (p < 0.005, n = 60 significant genes) in Atg12KO cells versus Atg12f/f cells in either fed or starved conditions.