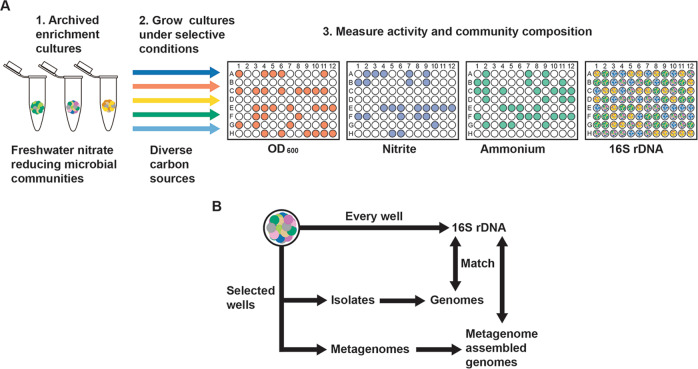
Fig. 1. Workflow to measure the influence of selective growth conditions on microbial community composition, gene content, and functional activity.
a Archived microbial enrichment cultures are cultured under different growth conditions. Community functional traits, community composition and both strain and community genetic potential are measured. In the present work, freshwater nitrate reducing microbial communities are grown on 94 different carbon sources, some of which are selective for different end products or intermediates of nitrate reduction. Growth (optical density/OD 600), nitrite and ammonium are measured through colorimetric assays, and microbial community composition is determined using 16S rDNA amplicon sequencing. b Pure culture microbial isolates, isolate genomes, and metagenome assembled genomes (MAGs) are obtained for select cultures. Matches between 16S rDNA sequences in genomes and MAGS with amplicons allows the assignment of genetic potential to the 16S rDNA exact sequence variants (ESVs) in all of the enrichment cultures.