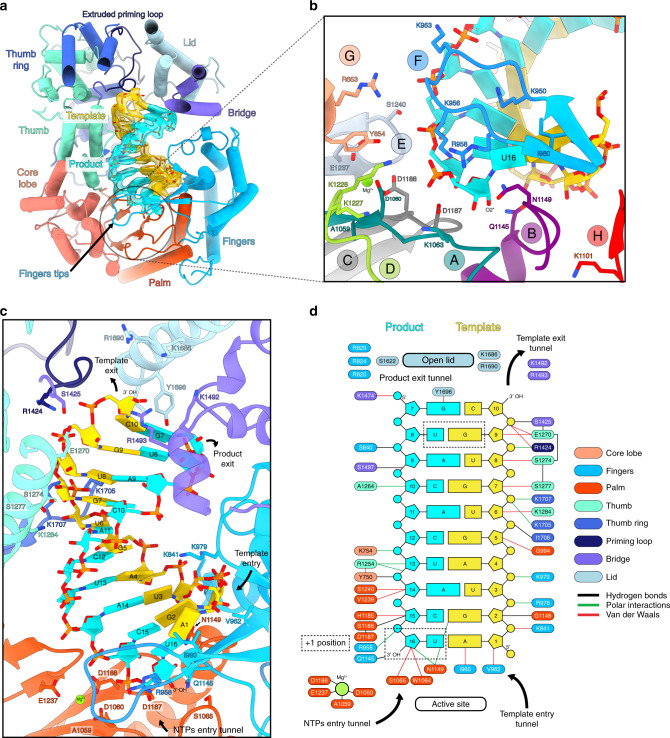
Fig. 5. Cryo-EM structure of the LACV-L FL at elongation-mimicking stage.
a Cut-away view of the LACV-L FL at elongation-mimicking stage. Its orientation corresponds to a top view visualization of Fig. 1b left. The main domains are depicted and colored as in Fig. 1. The Coulomb potential map of the 10-base-pair product–template RNA is shown in cyan for the product (3′OH-UCAUCACAUG, nucleotides 7–16) and gold for the template (5′p-AGUAGUGUGC, nucleotides 1–10). The extruded priming loop is shown in dark blue. The active site position is indicated as a dotted circle. b View of the LACV-L FL active site showing the conserved RNA-dependent RNA polymerase functional motifs A–H (G and H are only conserved in sNSV polymerases). They are respectively colored turquoise, purple, gray, light green, light blue, blue, beige, and red for A–H. Template-mimicking and product-mimicking RNA are colored as in a. Presumed magnesium ion, is shown as a green sphere. c Interactions of the 10-base-pair product–template RNA in LACV-L active site cavity. Principal residues from the active site in palm domain (A1059, D1060, S1065, D1187, D1188, E1237), fingertips (R958), fingers (K841, I960, V962, K979, Q1145), priming loop (R1424), bridge (S1425, K1492, R1493), thumb (E1270, S1274, S1277, K1284), and lid (K1686, R1690, Y1696) are displayed. NTPs entry, template entry, template entry/exit tunnel directions are shown. Ion position is shown as in b. Nucleotides are labeled according to RNA promoter sequence. d Schematic representation of RNA–protein contacts in the active site cavity. Residues are colored according the domain to which they belong. Template and product RNA are numbered according to their position in the 5′ end promoter (5′p-AGUAGUGUGC, nucleotides 1–10) and the 3′ end promoter (3′OH-UCAUCACAUG, nucleotides 7–16). The U-G mismatch that is due to the non-perfect complementarity between the 5′ and 3′ promoters is surrounded by a dotted rectangle. Interaction type are color coded as indicated. Ion is shown as a green circle. Active site and lid domain positions are indicated. Nucleotide U16 corresponds to the nucleotide in position +1 of the product and is identified as such.