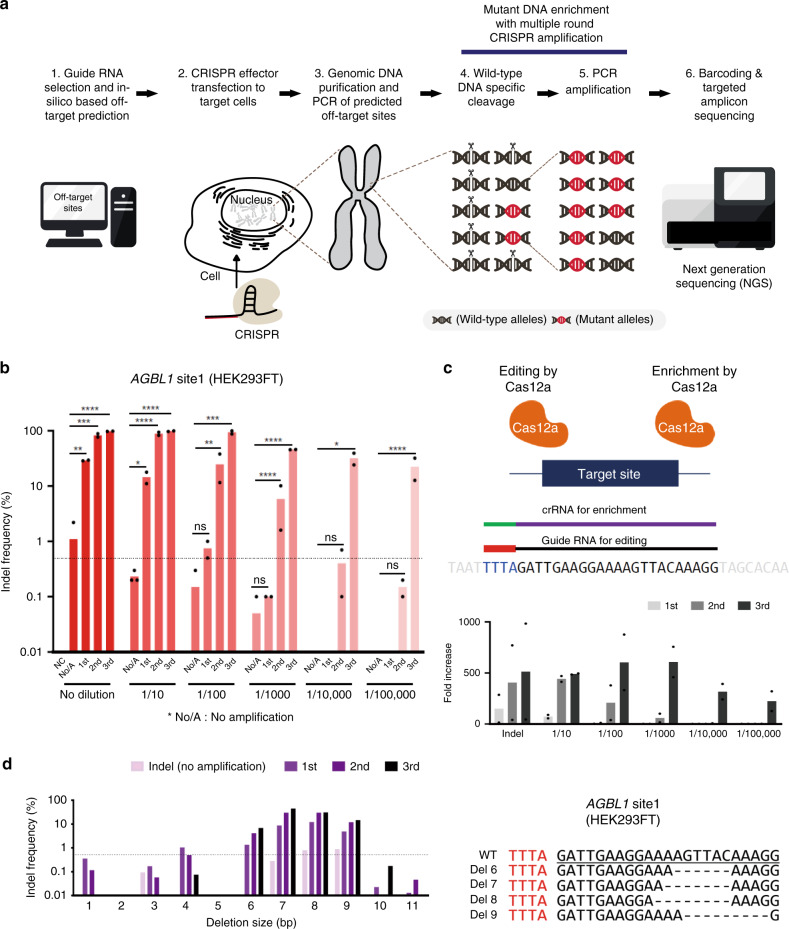
Fig. 1. Schematics of the detection of off-target mutations using the CRISPR amplification method.
a Workflow of the off-target detection method using CRISPR amplification. (1) Based on candidate off-target sequences predicted in silico, a gRNA is designed to enrich off-target sequences by subsequent CRISPR cleavage. (2) The CRISPR effector is transfected into cells to induce specific mutations. (3) Genomic DNA is extracted from the cells and predicted off-target loci are PCR-amplified. (4) The PCR amplicons are cleaved by the CRISPR effector. (5) DNA with non-cleaved indel mutations are amplified preferentially over wild-type DNA. (6) The amplified DNA is barcoded and analyzed by NGS. b Quantitative analysis of mutant DNA enrichment by CRISPR amplification. Genomic DNA samples with mutations induced in the target sequence were serially diluted (up to 1/100,000) and amplified. The indel efficiency (%) was calculated by sequencing DNA amplicons obtained from genomic DNA extracted from CRISPR–Cas12a edited cells. The X axis represents the degree of dilution of the genomic DNA samples, and the Y axis represents the indel detection rate (%). NC indicates the negative control of no genome editing. No/A indicates the genome-edited sample, but no amplification. The dashed line indicates the NGS detection limit (=0.5%). Data are shown as mean from two (N = 2) independent experiments. P-values are calculated using a one-way ANOVA, Tukey’s test (ns: not significant, *P = 0.0332, **P = 0.0021, ***P = 0.0002, ****P = 0.0001). c Top: design of the crRNA for editing and mutant DNA enrichment with Cas12a, respectively. crRNA was used to induce target genomic locus mutation by AsCas12a effector and exactly same crRNA was used for mutant DNA enrichment by CRISPR amplification. Bottom: fold increase of indel frequency by CRISPR amplification (N = 2) for each diluted samples from originally mutation-induced genomic DNA. d NGS analysis of AsCas12a-induced indel patterns enriched by CRISPR amplification. Left: gradual amplification (no amplification, primary, secondary, and tertiary amplification) of mutant DNAs with different deletion sizes. Each amplification stage is indicated by different colors. Right: representative enriched mutation patterns from third-round of CRISPR amplification with on-target amplicon. Source data is in the Source Data file.