Figure 5.
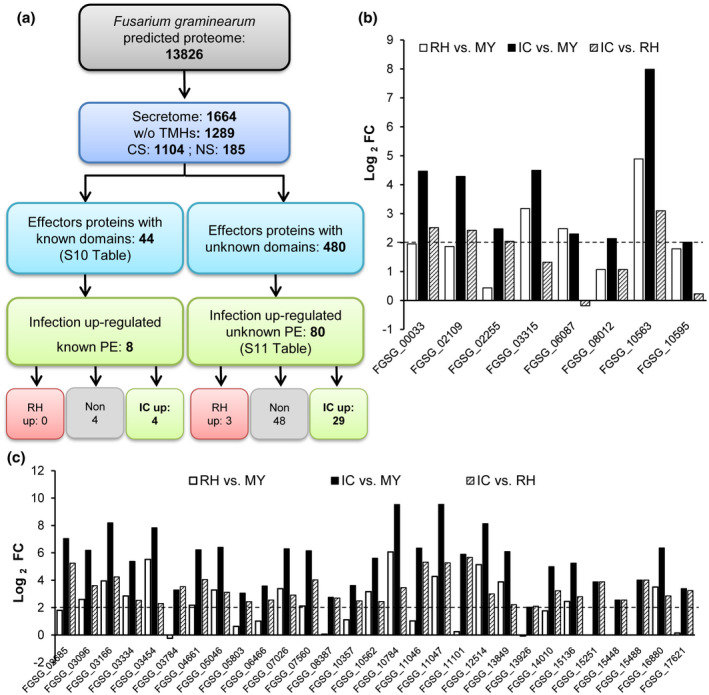
F usarium graminearum secretome prediction and effectors selection. (a) The secreted proteins were predicted using TargetP and SignalP for classic secretion (CS), or secretomeP and wolfPSORT for non‐classic secretion (NS). Proteins without transmembrane domains (w/o TMHs) were predicted using TMHMM. We identify putative effector proteins (PE) similar to effectors with known domains using IPRO or PFAM prediction domains (Table S10). Proteins with unknown domains, not belonging to a secondary metabolism gene cluster, with fewer than 1,000 amino acids, were defined as putative unknown effectors (Table S11). (b) Expression (log2 fold change [FC]) of the eight infection up‐regulated known PE. Four genes encode LysM proteins (FGSG_00033, FGSG_02255, FGSG_06087, and FGSG_10563), three for peptidase/proteinase inhibitor (FGSG_03315, FGSG_08012, and FGSG_10595), and one for CFEM protein (FGSG_02109). (c) Log2 FC of the 29 infection and IC up‐regulated unknown PE. Dashed lines show the log2‐threshold of +2 for up‐regulated genes. IC, infection cushions, RH, runner hyphae; MY, mycelium grown in complete medium
