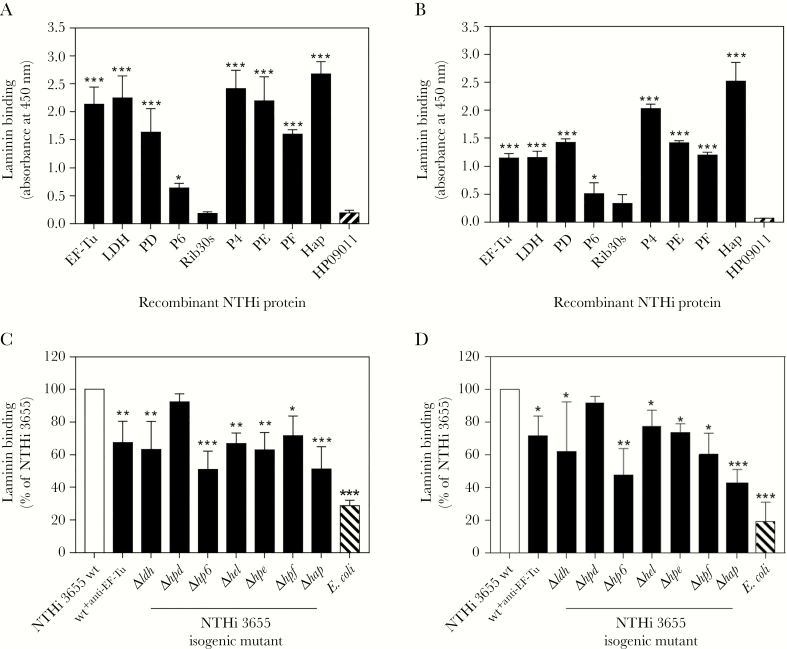
Figure 2.
Interaction of nontypeable Haemophilus influenzae (NTHi) novel laminin-binding proteins (Lbps) (elongation factor thermal unstable [EF-Tu], l-lactate dehydrogenase [LDH], protein D [PD], and lipoprotein P6 [P6]) with laminin as determined by enzyme-linked immunosorbent assay and a direct binding assay. (A) Binding of soluble laminin (10 nM) to immobilized His-tagged NTHi Lbps (EF-TuM1-K394, LDHM1-L381, PDS19-K364, P6S21-Y153, Rib30sM1-E125, P4G22-K274, PE22-160, PF12-293, and HapE523-L1036). Laminin binding was detected with rabbit antimouse laminin and horseradish peroxidase (HRP)-swine antirabbit polyclonal antibodies (pAbs). (B) Relative binding capacities of NTHi Lbps (as listed in A) (10 nM) to immobilized laminin. In this assay, binding of NTHi proteins to immobilized laminin were detected with an HRP-conjugated anti-His pAb. In A and B, His-tagged NTHi protein HP09011 (GenBank accession number EDJ93055) that does not bind laminin was included as negative control [9]. (C) Direct ligand-binding assay of NTHi 3655 wild-type (wt), NTHi 3655+anti-EF-Tu, and Lbp-deficient mutants with soluble laminin as analyzed by flow cytometry. The laminin deposition on wt and knockout mutants was detected with rabbit antimouse laminin pAb and fluorescein isothiocyanate (FITC)-conjugated swine anti-rabbit pAb. The laminin binding of NTHi 3655+anti-EF-Tu was separately analyzed with rat antimouse laminin β1 pAb (AbCam) and FITC-conjugated goat antirat immunoglobulin G pAb (AbCam). Escherichia coli Top10 that did not bind laminin was included as negative control [9]. Data are presented as percentage of bacterial binding to laminin relatively to the NTHi 3655 wt that was set as 100%. (D) Adherence of NTHi 3655 wt, NTHi 3655+anti-EF-Tu, and Lbp-deficient mutants to immobilized laminin as assayed by a colony counting method. Data are presented as percentage of bacteria colony-forming units retrieved from adhesion to immobilized laminin relatively to the NTHi 3655 wt that was set as 100%. In A–D, all data represent mean values of three independent experiments and standard deviations are indicated by error bars. Differences between recombinant Lbps and control protein HP09011 in A and B, and between wt and mutants or NTHi 3655+anti-EF-Tu in C and D, were calculated by one-way ANOVA. *, P ≤ .05; **, P ≤ .01; and ***, P ≤ .001.