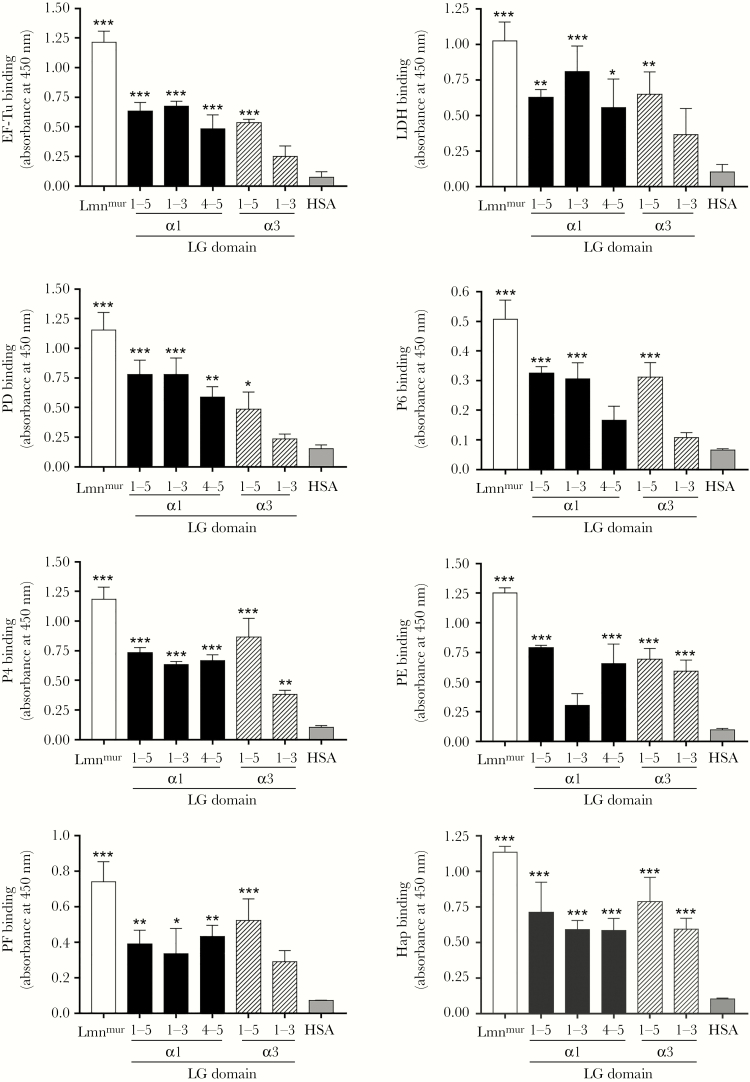
Figure 5.
Relative interactions of the recombinant nontypeable Haemophilus influenzae (NTHi) interactome with LG domains of laminin α1 and α3 chains as analyzed by enzyme-linked immunosorbent assay. Immobilized recombinant LG fragments of laminin α1 (LG1–5, LG1–3, and LG4–5) and laminin α3 (LG1–5 and LG1–3) were incubated with NTHi laminin-binding proteins (Lbps) (10 nM) (EF-TuM1-K394, LDHM1-L381, PDS19-K364, P6S21-Y153, P4G22-K274, PE22-160, PF12-293, and HapE523-L1036). Binding of NTHi proteins to the LG fragments were analyzed with Lbp-specific rabbit polyclonal antibody (pAb) (anti-EF-Tu [15], anti-LDH, anti-PD [16], anti-P4 [9], anti-PE [9], anti-PF [13], and anti-Hap [9]) and mouse anti-P6 monoclonal antibody (clone-7F3) [17], followed by horseradish peroxidase-conjugated swine antirabbit pAb or rabbit antimouse pAb as secondary antibodies. Lamininmur and human serum albumin (HSA) were included as positive and negative controls, respectively. Data represent mean values of three independent experiments, and error bars indicate standard deviations. Statistical differences between the binding of laminin or LG fragments, and HSA by each Lbp were calculated by one-way ANOVA. *, P ≤ .05; **, P ≤ .01; and ***, P ≤ .001. Lmnmur, lamininmur.