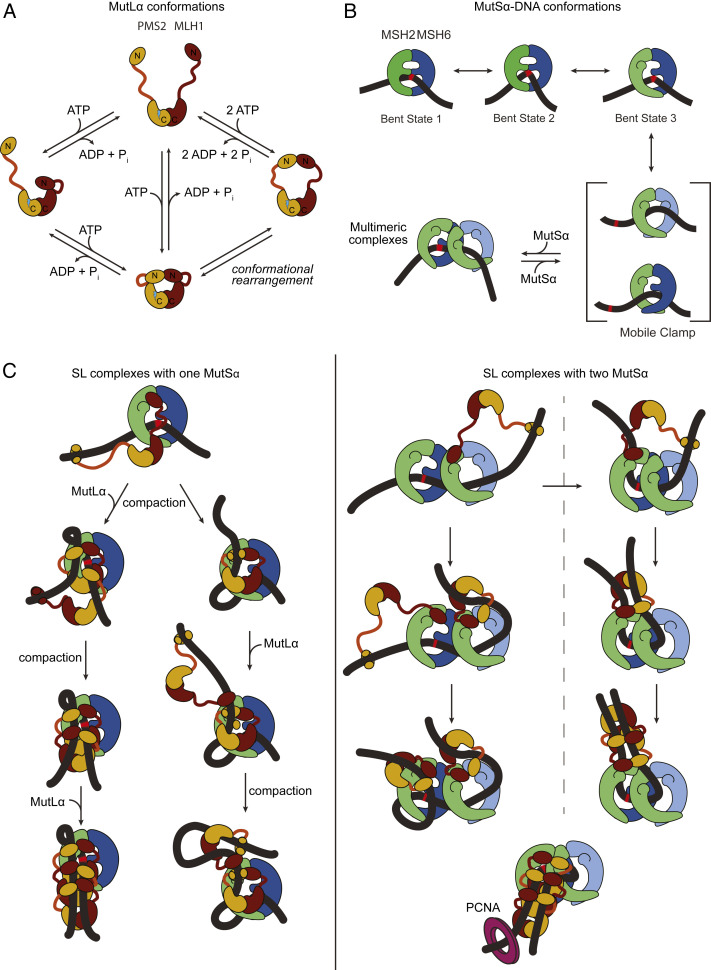
Fig. 5.
Example models of potential pathways to the formation of compacted MutSα–MutLα–GT-DNA complexes. (A) Previously identified conformational states of MutLα (42). MLH1 and PMS2 are shown in burgundy and ochre, respectively. The C-terminal dimerization domains are connected to the N-terminal ATPase and DNA binding domains by a flexible linker. The endonuclease site of PMS2 is shown as a lightning bolt. (B) Model for MutSα recognition, mobile clamp formation, and association of multiple MutSα. The early states (bent states 1 to 3) are from studies of Taq MutS (19), and the two mobile clamp conformations are based on the crystal structure of E. coli MutS mobile clamp in complex with the N-terminal domains of MutL (20), in conjunction with our recent single-molecule fluorescence studies that indicate that Taq MutS mobile clamps can exist in two conformations (87). (C) Example models for formation of SL complexes containing a single MutSα (Left) or two MutSα (Right). The Discussion includes a detailed description. Complexes are shown at the mismatch, but they can also occur at nonspecific sites on GT-DNA. These models also suggest that loop formation and DNA compaction could both result from the MutLα N-terminal domain binding distally on the DNA with one of its arms in an extended state (as in A), followed by nucleotide-induced retraction of that arm toward the C-terminal domain containing the endonuclease site. The lower right model depicts how PCNA (purple ring) could interact with an SL complex with antiparallel DNA strands to activate MutLα to nick on either side of the mismatch depending on the orientation of MutLα (15). The conformations of MutSα and MutLα depicted in the model are based on the conformations shown in A and B, and the location of the interactions between MutSα and MutLα are derived from the crystal structure of E. coli MutS sliding clamp in complex with the N-terminal domains of MutL (20) and from hydrogen/deuterium exchange mass spectrometry studies of E. coli MutS and MutL coupled with functional studies of yeast MutSα and MutLα (92). Models are only representative and are not intended to imply specific pathways or any structural details, but rather to provide ideas of how MutSα–MutLα complexes could compact the DNA.