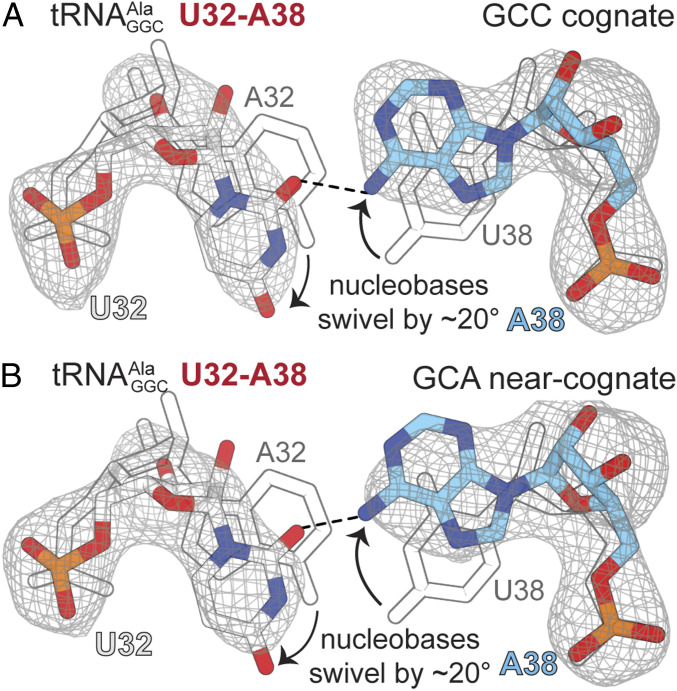
Fig. 5.
Reversing the 32–38 pair in causes both nucleotides to swivel ∼20° in opposite directions, disrupting canonical Watson–Crick base pairing. (A) The structure of 70S U32−A38 interacting with a cognate GCC codon. (B) The 70S U32-A38 variant interacting with a near-cognate GCA codon. In both structures, the nucleotides swivel away from each other, as compared to the position of the A32−U38 base pair in wild-type tRNA bound to the cognate codon (shown in gray outline). The 2FO−FC electron density maps in gray are contoured at 1.0σ.