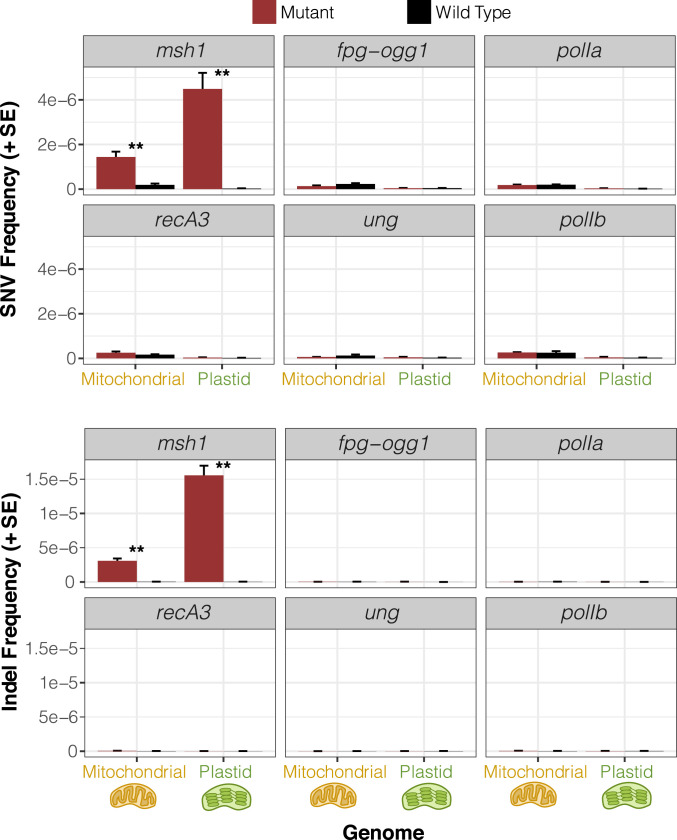
Fig. 4.
Observed frequency of mitochondrial and plastid SNVs (Top) and indels (Bottom) based on duplex sequencing in Arabidopsis mutant backgrounds for various RRR genes compared to matched wild-type controls. Variant frequencies are calculated as the total number of observed mismatches or indels in mapped duplex consensus sequences divided by the total base pairs of sequence coverage. Means and SEs are based on three replicate F3 families for each genotype (Fig. 2). The msh1 mutant line reported in this figure is CS3246. **Significant differences between mutant and wild-type genotypes at a level of P < 0.01 (t tests on log-transformed values). All other comparisons were nonsignificant (P > 0.05).