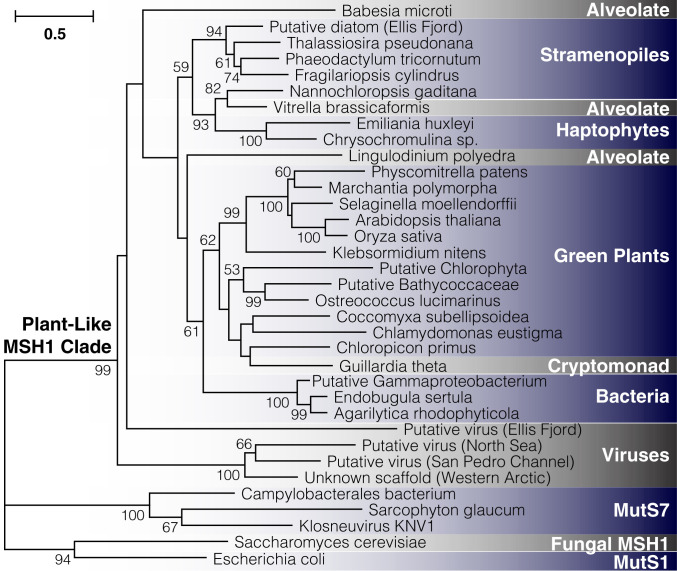
Fig. 7.
Detection of plant-like MSH1 genes across diverse evolutionary lineages. The maximum-likelihood tree is constructed based on aligned protein sequences, with branch lengths indicating the number of amino acid substitutions per site. Support values are percentages based on 1,000 bootstrap pseudoreplicates (only values >50% are shown). Metagenomic samples are putatively classified based on other genes present on the same assembled contig (SI Appendix, Table S6, provides information on sequence sources). The sample labeled “unknown scaffold” lacks additional genes for classification purposes, but it clustered strongly with two sequences from the IMG/VR repository of viral genomic sequences.