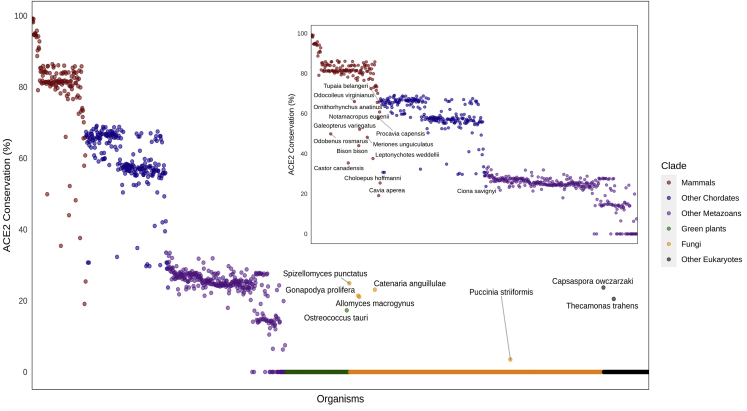
Figure 1.
Evolutionary Pattern of ACE2 Proteins across 1,671 Species
The percentage of sequence similarity (y axis) to the human ACE2 protein when compared with the most similar orthologs across all eukaryotes (x axis) is presented. The Conservation score is defined as Pab/Paa, where Pab is the best BLASTP bit-score between a Homo sapiens protein “a” and all open reading frames of a eukaryote genome “b.” Paa is marked as the self-similarity score of the H. sapiens protein “a” when blasted against itself. Organisms with scores that deviated from their clade-average conservation rate are highlighted with the corresponding name. See also Figures S1 and S2, and Table S1.