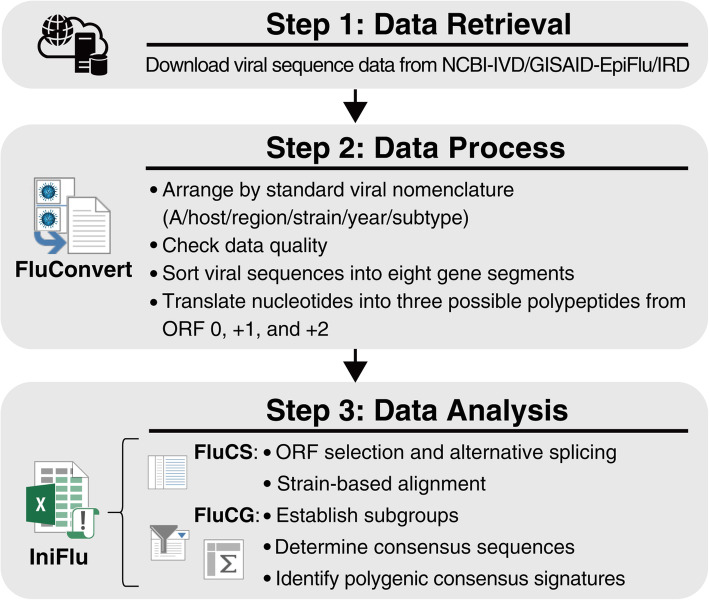
Fig. 1.
Workflows of data analysis executed by FluConvert and IniFlu. The stepwise processes performed by FluConvert and IniFlu to identify novel signatures of emerging influenza viruses with increasing risk are described as follows. Step 1: Viral sequences are obtained from the three databases (NCBI-IVD, GISAID-EpiFlu, and IRD). Step 2: FluConvert rearranges viral strains by viral nomenclature and ensures data quality. Viral sequences are further sorted into eight gene segments and translated into amino acid sequences. Step 3: The module FluCS of IniFlu performs strain-based alignments of FluConvert-processed viral amino sequences. The module FluCG of IniFlu regroups viral strains with epidemiological significant and computes a consensus sequence for each subgroup. Finally, the subgroup-specific unique polygenic amino acid signatures can be simultaneously identified (see details in Fig. 3)