Figure 1.
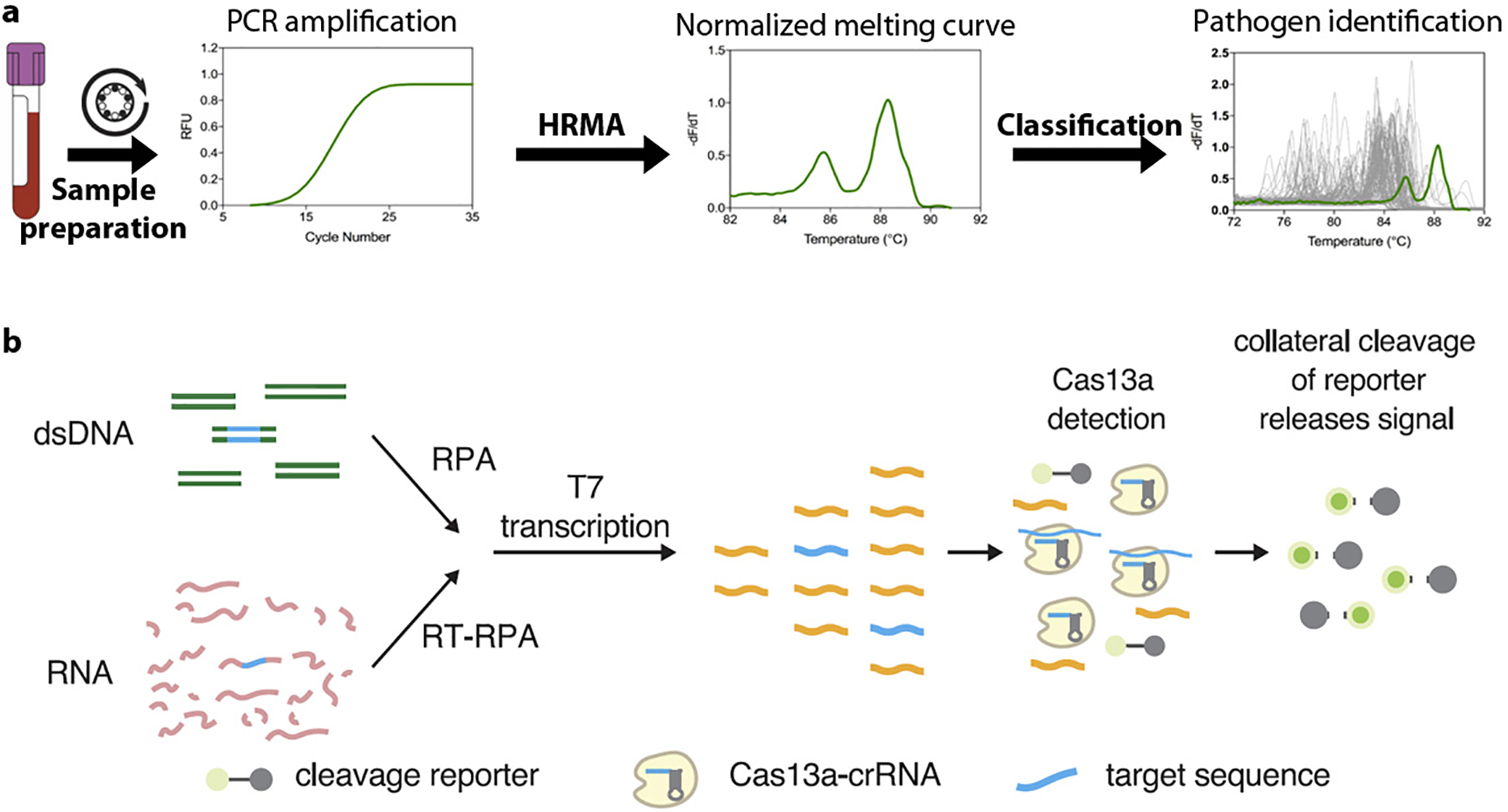
Emerging demonstrations of NAAT-based pathogen identification. (a) With HRMA, species were identified by their unique melting curves. After PCR amplification and HRMA, the raw melting curve of an unknown species was normalized and transformed into a derivative curve. An adaptive algorithm was then matched the unknown curve (green line) against an archived melting curve database (gray lines) to find the best fit. (b) Schematic of the SHERLOCK platform. The DNA or RNA target extracted from biological samples is amplified by RPA (RT-RPA or RPA, respectively). RPA products are detected in a reaction mixture containing T7 RNA polymerase, Cas13, a target-specific crRNA, and an RNA reporter that fluoresces when cleaved. Figure adapted with permission from Reference 48. Copyright 2017, AAAS. Abbreviations: crRNA, CRISPR RNA; HRMA, high-resolution melting analysis; NAAT, nucleic acid amplification technology; PCR, polymerase chain reaction; RFU, reflective fluorescence unit; RT, reverse transcription; RPA, recombinase polymerase amplification; SHERLOCK, specific high-sensitivity enzymatic reporter unlocking.
