Figure 7. Polymer modeling recapitulates key aspects of E. coli chromosomal structure.
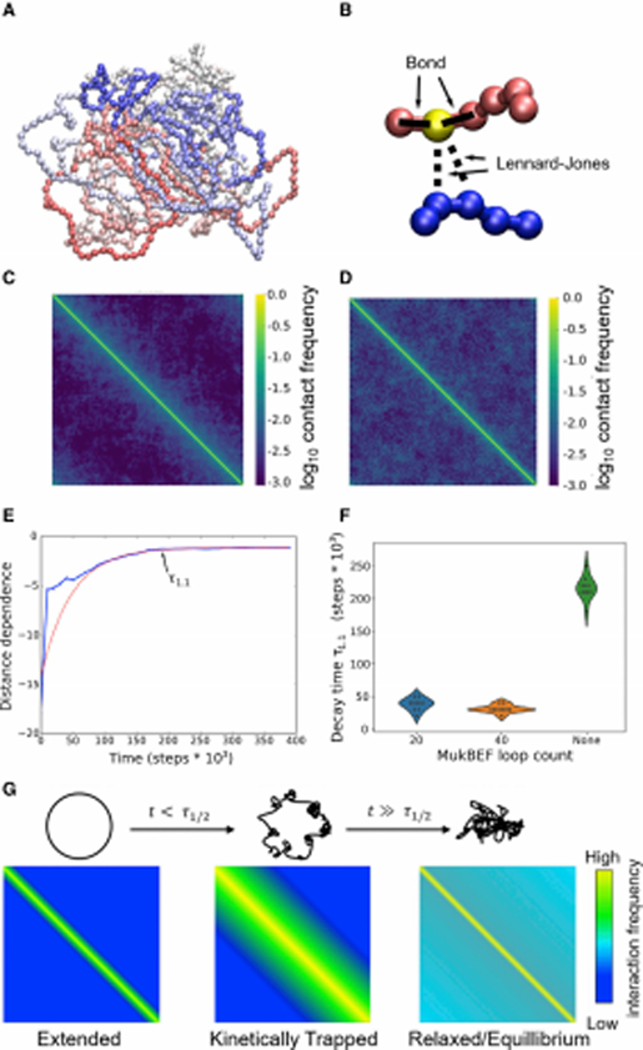
(A) Example conformation of the model at apparent equilibrium after relaxation from a fully extended ring shape. Coloring runs from blue to red showing linear position along the genome. (B) Schematic of two distant regions of the model chromosome and the interactions acting upon the yellow highlighted bead. (C) Contact matrix obtained by averaging contact maps from 50 short (106 timestep) simulations; see STAR Methods. (D) Contact matrix obtained by averaging the last halves of 20 long (2.5 * 107 timestep) simulations, showing the eventual equilibrium state reached by the system as it relaxes. Note that in all contact maps, the strong yellow stripe along the diagonal represents self-contacts (which thus have a frequency of 1.0 by definition). (E) Change in the dependence of contact frequencies from a representative polymer simulation over time. Distance dependences are obtained at each timepoint by fitting a logistic regression to predict contact frequency as a function of log(genomic distance); thus, a more negative distance dependence indicates stronger effects of linear distance. The red dashed line shows a single exponential fit to the observed decay curve beginning after 100,000 steps. T1.1 indicates the time needed to reach within 10% of the asymptotic value. (F) Violin plots showing the distributions of relaxation times (T1.1) for the distance dependence (as shown in panel E) in the absence vs. presence of MukBEF-induced loop extrusion. (G) Model for the progression of chromosomal states and contact maps in the polymer model. Schematic of the chromosomal conformation (top) and corresponding contact matrices (bottom) at the beginning of the simulations (left), after an amount of time shorter than the required relaxation time (middle), or after sufficient time for the conformation to reach equilibrium (right).
