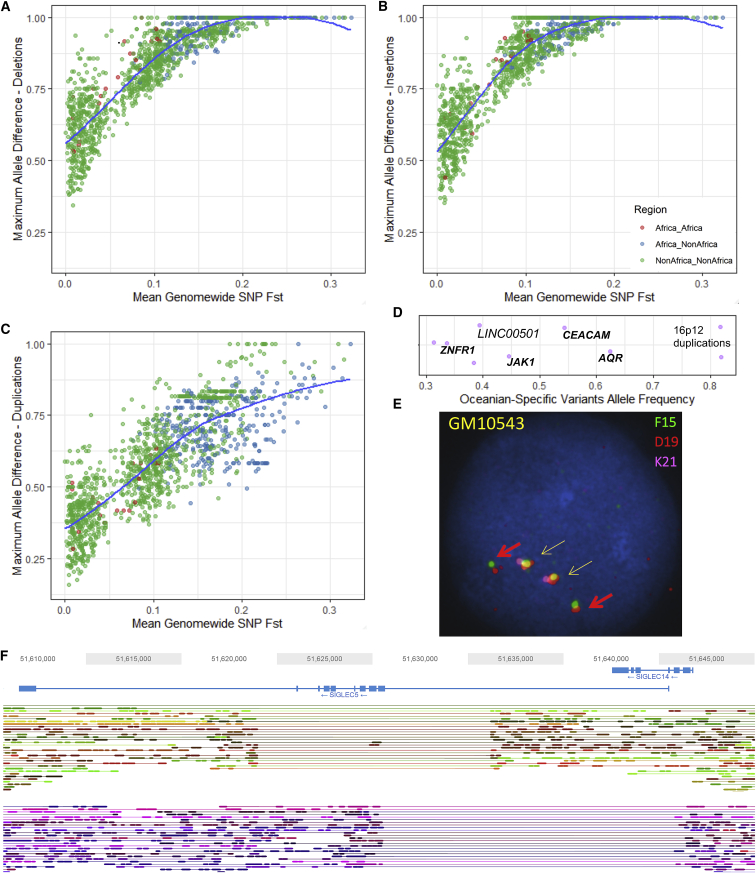
Figure 2.
Population Stratification of Structural Variants
(A) Maximum allele frequency difference of deletions as a function of population differentiation for 1,431 pairwise population comparisons. The blue curve represents locally estimated scatterplot smoothing (LOESS) fits.
(B and C) The same as (A) but for insertion (B) and biallelic duplications (C).
(D) High-frequency Oceania-specific variants (>30% frequency). See Figure S3 for more details. Each point represents a variant, with the x axis illustrating its frequency. Random noise is added to aid visualization. Almost all variants are shared with the Denisovan genome and are within (bold) or near the illustrated genes.
(E) Fluorescence in situ hybridization illustrating the 16p12 Oceania-specific duplication shared with Denisova in a homozygous state (cell line GM10543). Yellow arrows show reference, and red arrows illustrate duplication. See Figure S6 and S7 for more details.
(F) Distinct deletions at the SIGLEC5/SIGLEC14 locus in an Mbuti sample (HGDP00450), resolved using linked reads. Lines connecting reads illustrate that they are linked; i.e., they are from the same input DNA molecule. One haplotype (top) carries the Mbuti-specific variant that deletes most exons in SIGLEC5 and is present at high frequency (54%), whereas the second haplotype (bottom) carries a globally common deletion that deletes SIGLEC14, creating a fused gene (see STAR Methods for more details).