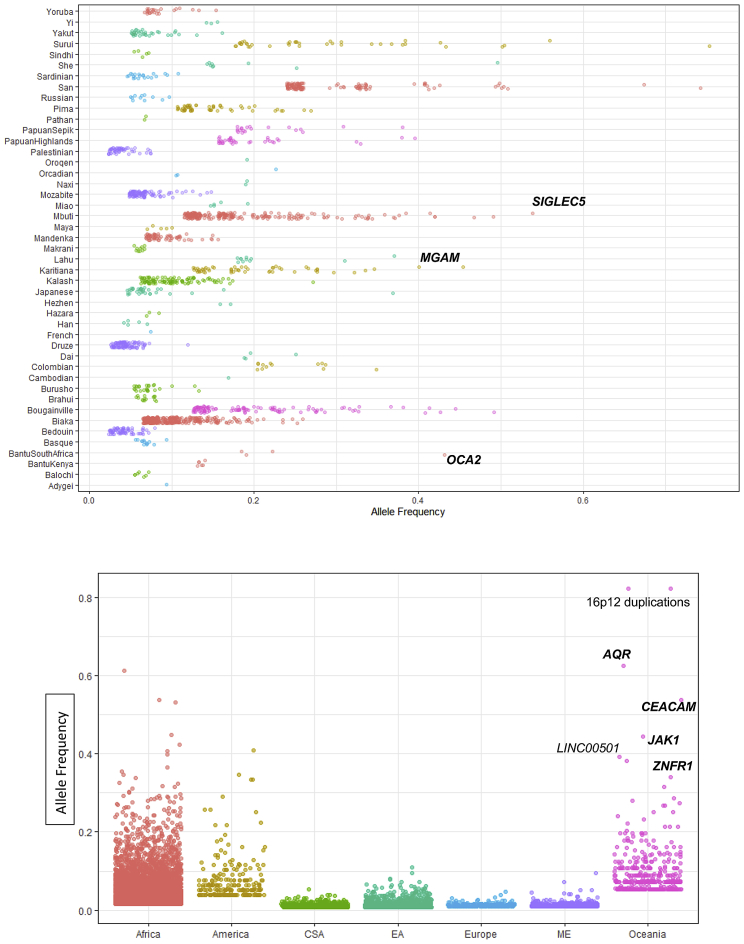
Figure S3.
Population- and Region-Specific Variation, Related to Figures 2D–2F
Top: Population-specific variation - Each point represents a variant private to a population (n > 2) with the x axis reflecting its frequency. Colors represent regional labels and random noise is added to aid visualization. High-frequency variants discussed in the text are highlighted. Bottom: Regional-Specific Variation – Each point represents a variant private to a regional group (n > 2) with the y axis illustrating its frequency. Random noise is added to aid visualization. The distribution reflects the ancestral diversity in Africa, the connectivity of Eurasia, the isolation & drift of the Americas and Oceania, and the separate Denisovan introgression event in Oceania. Oceania is notable for having private high-frequency variants that are all shared with the Denisovan genome and are within (bold) or near the illustrated genes, four of which are newly identified in this study (AQR, CEACAM, JAK1, ZNFR1). The Americas contain high frequency variants which are not shared with any archaic genomes, suggesting they arose and increased to high-frequency after they split from other populations. EA: East Asia, CSA: Central & South Asia, ME: Middle East.