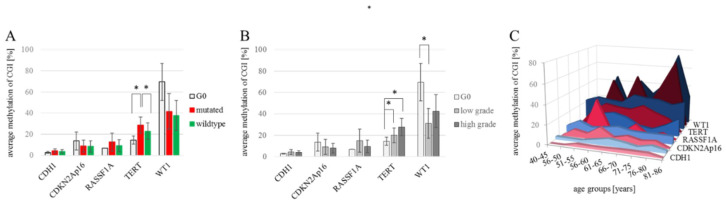
Figure 1.
Developments of CDH1/CDKN2Ap16/RASSF1A/TERT/WT1 methylation levels. (A) Average promoter methylation levels for each gene of interest for tumor-free control samples (nCDH1 = 2; nCDKN2Ap16 = 7; nRASSF1A = 1; nTERT = 7; nWT1 = 3), wildtype (nCDH1 = 34; nCDKN2Ap16 = 89; nRASSF1A = 22; nTERT = 78; nWT1 = 23) and EGFR/KRAS mutated tumor samples (nCDH1 = 22; nCDKN2Ap16 = 46; nRASSF1A = 19; nTERT = 39; nWT1 = 16). Significant changes are measured for TERT methylation; i.e., G0 vs. mutated, p = 0.050 and mutated vs. wild-type p = 0.046. Student’s t-test. (B) Average methylation of each gene in non-tumor/G0, low grade (G1-G2) (nCDH1 = 14; nCDKN2Ap16 = 43; nRASSF1A = 11; nTERT = 33; nWT1 = 18), and high grade tumor samples (G3-G4) (nCDH1 = 42; nCDKN2Ap16 = 92; nRASSF1A = 30; nTERT = 85; nWT1 = 21). A statistically significant increase of TERT methylation was found comparing tumor-free control to low-grade tumors (p = 0.022) and to high grade tumors (p = 0.021), the steady increase with tumor progression is a significant correlation (p = 0.003) Spearman correlation. WT1 methylation decreased significantly comparing tumor-free control to low grade tumor samples (p = 0.043) Student’s t-test. (C) Methylation of target genes in the patient samples grouped by gender and age. A Spearman correlation analysis matrix showed no significant correlation between overall age and promoter methylation for any of the analyzed genes. * p < 0.05.