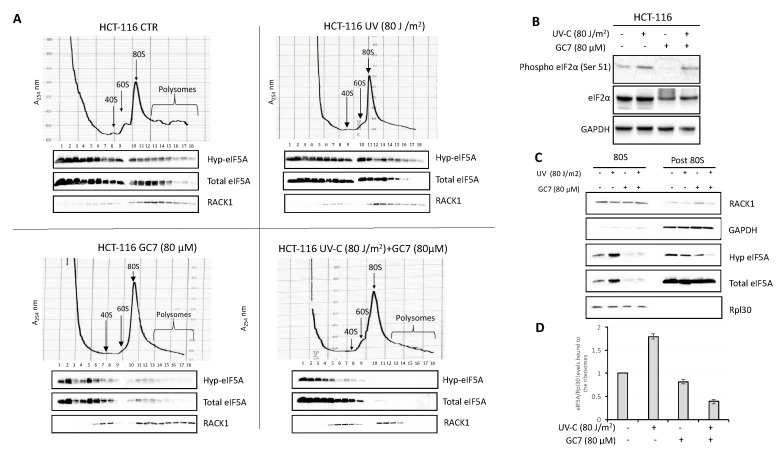
Figure 5.
GC7 and UV treatments affect the binding of eIF5A to the ribosomes of HCT-116 cells. (A) HCT-116 cells polysomes profile was analyzed by density gradient centrifugation. After the indicated treatments, 10 ODs of the cell lysates were loaded onto a sucrose gradient and fractionated. Optical scans (OD254 nm) of the gradients are shown. Fractions were analyzed by immunoblotting with the indicated antibodies. (B) HCT-116 cells were treated with the indicated doses of UV-C and GC7. Whole cell extracts were prepared and phosphorylation and protein levels were determined by immunoblotting with the appropriate antibodies, as indicated. (C) Ribosomes (80S) were purified from HCT-116 cells by sedimentation through a sucrose cushion and were probed with the indicated antibodies. The post 80S represents the supernatant after ribosomes precipitation. The absence of Rpl30 protein shows the ribosome-free fraction. (D) Intensity of bands was detected by the Bio-rad Image Lab Software 5.2.1 and the signal of interest was normalized to control Rpl30. The value of eIF5A in untreated cells was set as 1. Data are ± SD (n = 3).