Figure 1.
Small Molecule Screen to Identify Enhancers of Antigen Import into the Cytosol
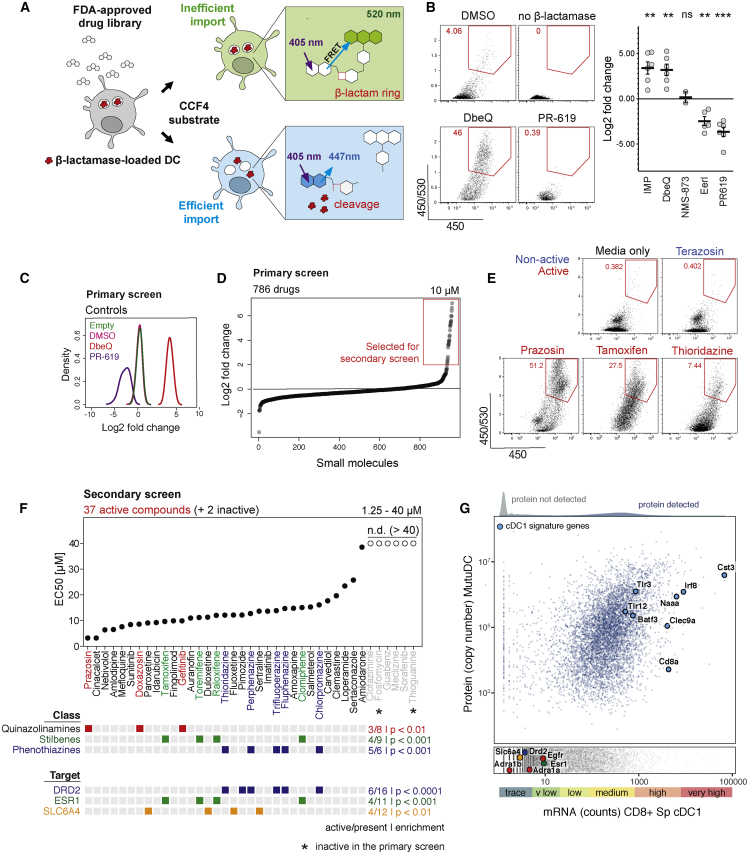
(A) Schematic representation of the β-lactamase assay used to monitor the efficiency of antigen import into the cytosol. MutuDCs were fed with β-lactamase for 3 h followed by 2 h incubation with small molecules (at 37°C). CCF4 loading was performed at room temperature for 1 h, and followed by 16 h incubation at RT to increase the sensitivity of the assay (Zlokarnik et al., 1998). Change in CCF4 fluorescence was monitored by flow cytometry.
(B) Differential effects of ERAD inhibitors on antigen import into the cytosol. Representative flow cytometry data for selected ERAD inhibitors and quantification of the fold change in antigen import relative to DMSO controls. IMP, importazole; EerI, Eeyarestatin I. Means ± SE (dots represents data from independent experiments).
(C) Quality control of the FDA library screen. The histograms show distribution of fold changes in the efficiency of antigen import (relative to DMSO) for each control (all wells across the ten 96-well plates).
(D) Results from the FDA library screen. Fold changes in β-lactamase import for the 786 tested drugs. The screen was performed once, and 37 compounds were selected for the secondary screen (highlighted with the red box).
(E) Examples of the flow cytometry profiles in the antigen import assay for selected active and non-active compounds.
(F) Results from the secondary screen of 37 compounds (and two control compounds, not active in the primary screen). Each drug was tested at five concentrations in two independent experiments. EC50 values (concentration required for 50% of maximal activity) values were calculates as described in Figure S2. Information about chemical classes and candidate targets was obtained from the DrugBank database. The classes and targets enriched in the group of active versus non-active compounds are represented with colored squares. The enrichment of targets for hits (compared to the entire library) was calculated using Fisher’s exact test.
(G) Analysis of gene and protein expression in CD8+ cDC1s. mRNA expression data (RNA sequencing [RNA-seq]) for CD8+ splenic DCs was downloaded from the www.Immgen.org database (GEO: GSE109125), and whole cell proteomic abundance data were generated by mass spectrometry from MutuDCs. 7427 proteins were detected by proteomics (blue dots) and selected markers highly expressed in cDC1s are highlighted with large blue circles. Absolute copy numbers for all proteins detected in whole cell MutuDC proteome are available via the web resource (http://dc-biology.mrc-lmb.cam.ac.uk). The lower panel (gray dots) includes proteins not detected by proteomics. Targets enriched in the group of active versus non-active compounds are highlighted (Esr1, Drd2, and Slc6a4), as well as targets of the three active quinazolinamines (Adra1b for prazosin, Adra1a for doxazosin, and Egfr for gefitinib).
See also Figures S1 and S2 and Data S1 and S2.