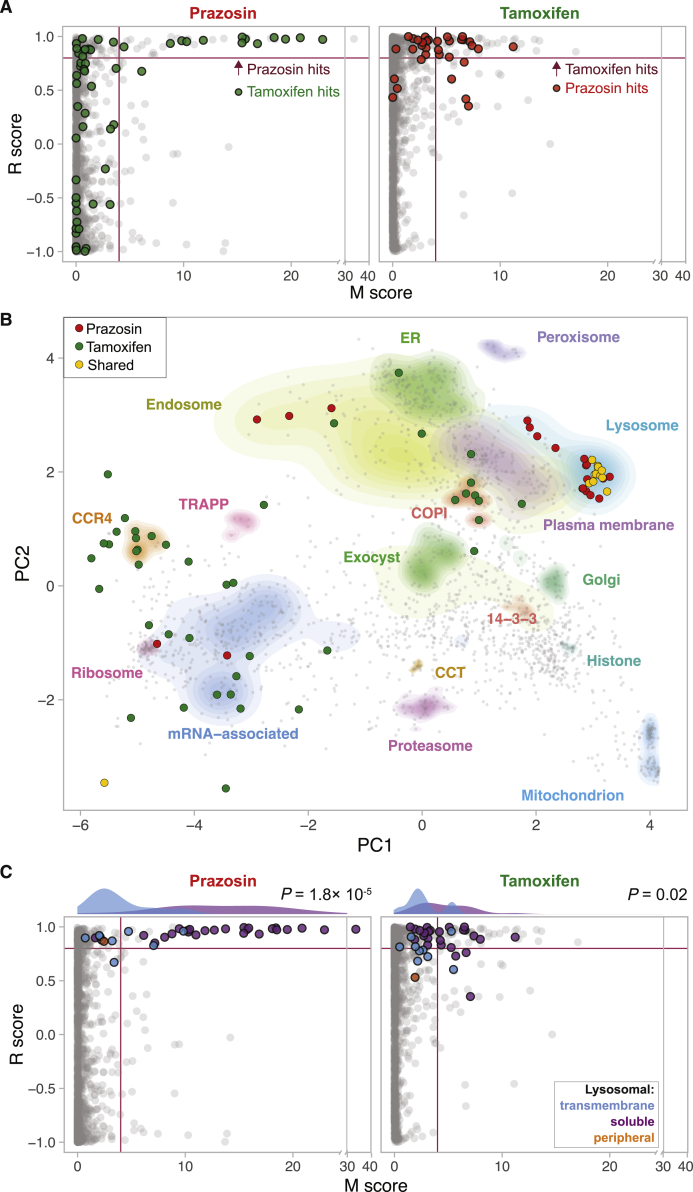
Figure 3.
Dynamic Organellar Mapping to Identify the Subcellular Changes in Protein Distribution on Drug Treatment
MutuDCs were treated with either prazosin, tamoxifen, or DMSO (control) for 4 h in biological duplicate, and samples were processed as described in Figure 2A. Statistical comparison of organellar maps made with different treatments was performed to identify proteins with profile shifts/altered subcellular localization.
(A) Drug-induced shifts in protein subcellular localization detected using a “MR” plot analysis. For each protein, the movement (M score) and the reproducibility of the movement (R score) between maps was determined. Purple lines indicate cut-offs for significance. In the prazosin plot the hits from tamoxifen treatment are shown for comparison (and vice-versa). Most prazosin hits are also tamoxifen hits or near-hits.
(B) Shifting proteins from tamoxifen and prazosin-treated samples represented on the organellar map of MutuDCs. Most shared hits are lysosomal proteins.
(C) MR plot highlighting all detected lysosomal proteins (soluble, transmembrane, and peripheral [located on the cytosolic side of the membrane]). The histograms show distribution of the M scores for transmembrane and soluble lysosomal proteins.
p values were calculated using the Mann-Whitney U test. See also Figure S3 and Data S2.