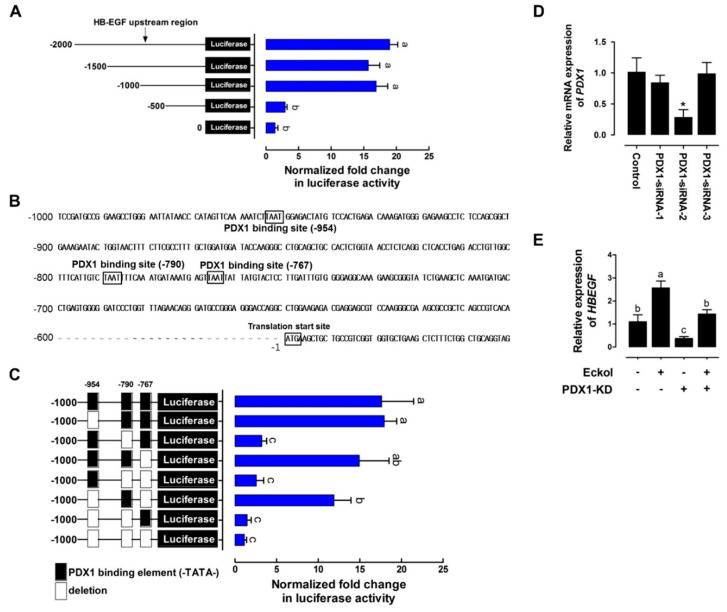
Figure 4.
Eckol-induced PDX1 regulates HBEGF expression. (A) Activity of HBEGF upstream region. IPEC-J2 cells were transfected with DNA sequences of different lengths (−2000, −1500, −1000, and −500) comprising the upstream region of HBEGF (n = 3). (B) Upstream sequence of the putative core region of the HBEGF promoter. Sequence numbering is relative to the transcription start site. Putative PDX1 binding sites (TAAT) are boxed and labeled above (upstream of −954, −790, and −767). (C) Deletion assay of putative PDX1 binding sites. Sequences in which the deleted binding sites (upstream of −954, −790, and −767) were transfected into IPEC-J2 cells (n = 3). (D) PDX1 knockdown assay. The siRNA-mediated suppression of PDX1 in IPEC-J2 cells was confirmed using qRT-PCR. (E) Relative HBEGF expression in PDX1-silenced IPEC-J2 cells treated with eckol. Relative luciferase activity was calculated as the ratio of firefly to Renilla luciferase, and the relative expression level was normalized to that of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Error bars indicate standard error of triplicate analyses. * p < 0.05. Lower-case letters indicate significant differences among treatments based on Duncan’s multiple range test.