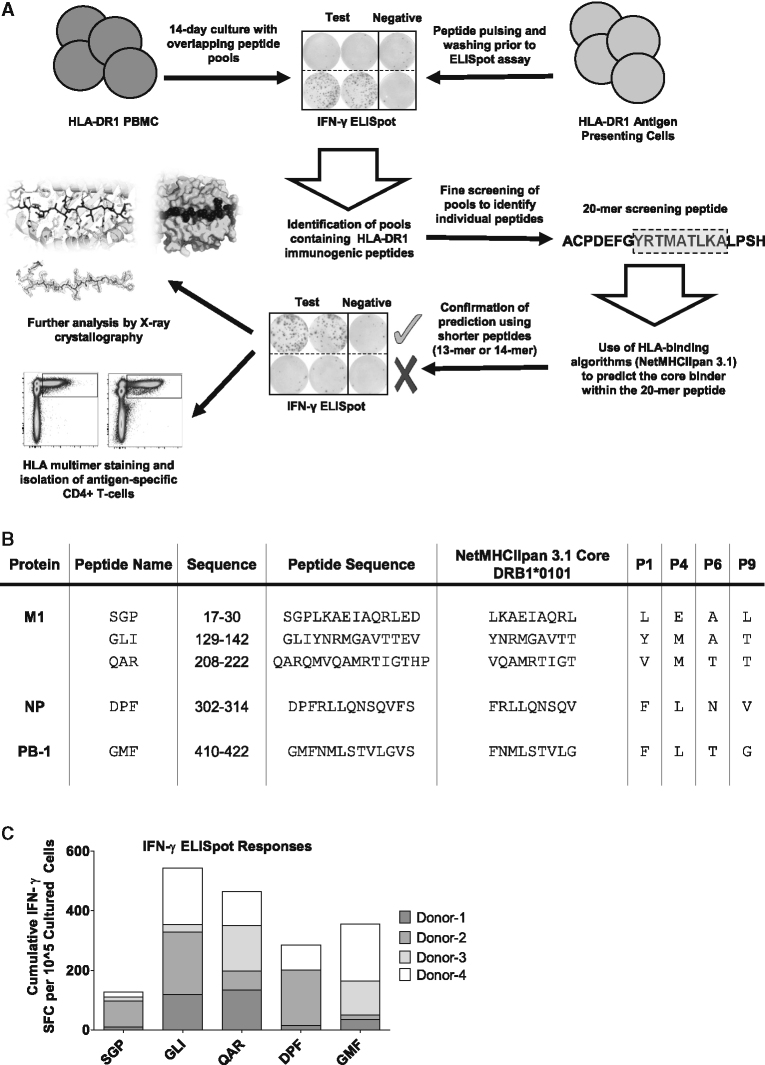
Figure 1.
Identification of HLA-DR1 Epitopes from Three Internal Proteins of IAV
(A) Schematic representation of epitope mapping procedure. HLA-DR1+ donor peripheral blood mononuclear cells (PBMCs) cultured with influenza peptide pools were screened on IFN-γ ELISpot by using peptide-pulsed HLA-DR1+ antigen-presenting cells (APCs), followed by identification of immunogenic peptides and use of NetMHCIIpan to elucidate the 9-amino-acid core. Shorter peptides were tested on IFN-γ ELISpot, followed by further validation and analysis using HLA-multimer screens and X-ray crystallographic analysis of peptide-HLA structures.
(B) Table of identified HLA-DR1 epitopes and final peptide sequences used for further analysis. Anchor residues P1, P4, P6, and P9 are listed in far-right column, as indicated by NetMHCIIpan.
(C) Cumulative IFN-γ ELISpot responses to identified peptides in four HLA-DR1+ donors. Responses to each peptide per donor (mean of two replicates per donor) were stacked to give the cumulative response in terms of SFC per 105 cultured cells.