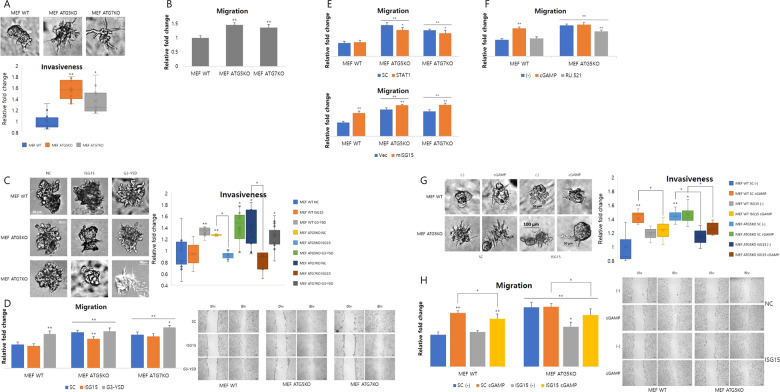
Fig. 7. ISG15 induction mediated by the STING pathway positively regulates tumorigenesis in Atg5- or Atg7-depleted cells.
a Invasiveness of indicated cells was identified using a 3D culture assay. Relative fold change of invasiveness is measured by Fiji and normalized to wild type (WT) (n = 3). *p < 0.05, **p < 0.01 (Student’s t test). b Migration of indicated cells was identified using a wound-healing assay. Relative fold change of migration is measured by Fiji and normalized to WT (n = 4). *p < 0.05, **p < 0.01 (Student’s t test). c After transfecting cells with the indicated siRNAs (50 pmol) or G3-YSD (10 μg/ml), invasiveness of the indicated cells was identified using a 3D culture assay. Relative fold change of invasiveness is measured by Fiji and normalized to WT (n = 3). *p < 0.05, **p < 0.01 (Student’s t test). d After transfecting cells with the indicated siRNAs (50 pmol) or G3-YSD (10 μg/ml), migration of the indicated cells was identified using a wound-healing assay. Relative fold change of migration is measured by Fiji and normalized to WT (n = 4). *p < 0.05, **p < 0.01 (Student’s t test). After treating cells with the indicated siRNAs (e), or drugs (f), migration of indicated cells was identified using a wound-healing assay. Relative fold change of migration is measured by Fiji and normalized to WT (n = 4). *p < 0.05, **p < 0.01 (Student’s t test). g After transfecting cells with the indicated siRNAs (50 pmol) followed by 2′3′′-cGAMP treatment, invasiveness of the indicated cells was identified using a 3D culture assay. Relative fold change of invasiveness is measured by Fiji and normalized to WT (n = 3). *p < 0.05, **p < 0.01 (Student’s t test). f After transfecting cells with the indicated siRNAs (50 pmol) followed by 2′3′-cGAMP treatment or not, migration of the indicated cells was identified using a wound-healing assay. Relative fold change of migration was measured by Fiji and normalized to WT (n = 4). *p < 0.05, **p < 0.01 (Student’s t test).