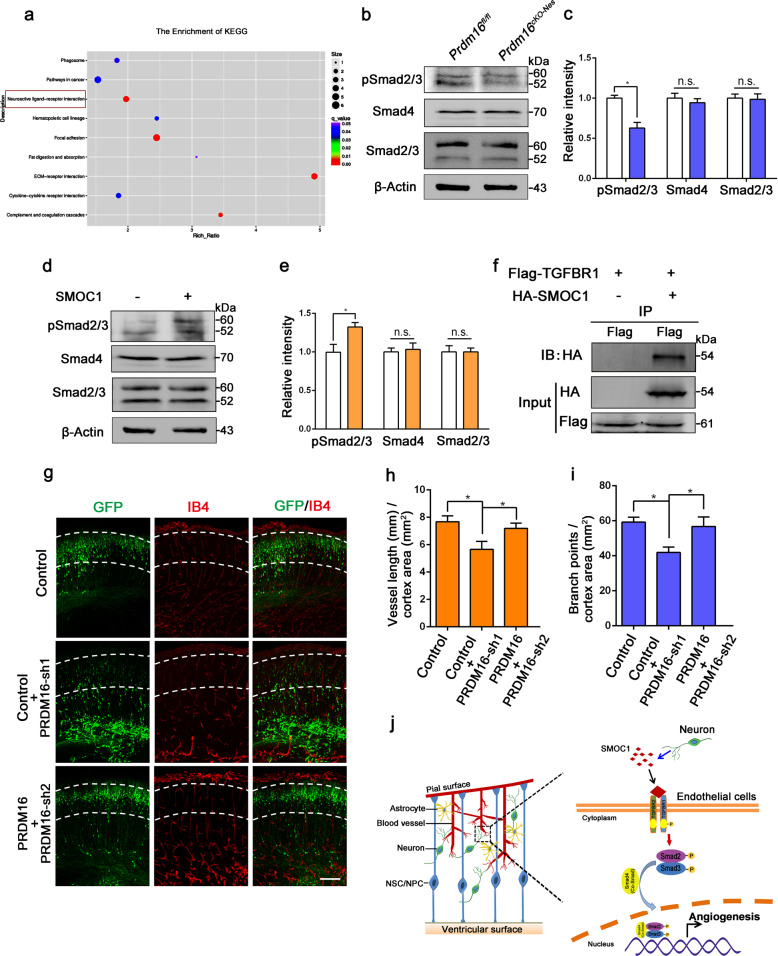
Fig. 6. PRDM16 regulates angiogenesis through the activation of the vascular TGF-β signaling pathway by SMOC1 during cortical development.
a Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was used to evaluate enriched pathways. The neuroactive ligand–receptor interaction pathway was dramatically enriched (red rectangle). b Western blot analysis of the expression levels of pSmad2/3, Smad4, and Smad2/3 in Prdm16fl/fl and Prdm16cKO-Nes cortices at E18.5. c Quantification of pSmad2/3, Smad4, and Smad2/3 expression levels in Prdm16fl/fl and Prdm16cKO-Nes cortices. The data are represented as the means ± SEM (n = 3; unpaired Student’s t test; *P < 0.05; ns not significant). d Western blot analysis of ECs with or without SMOC1 (10 ng/ml) stimulation. The expression levels of phosphorylated Smad2/3 were increased upon SMOC1 stimulation. e Quantification of pSmad2/3, Smad4, and Smad2/3 expression levels in SMOC1 stimulated ECs. The data are represented as the means ± SEM (n = 3; unpaired Student’s t test; *P < 0.05; ns not significant). f Western blotting of forward co-immunoprecipitation of SMOC1 and TGFBR1. g The disruption of angiogenesis caused by Prdm16 knockdown was partly rescued by PRDM16. A Prdm16 shRNA plasmid was coelectroporated with a PRDM16-expressing plasmid at E13.5, and then the brains were isolated at E18.5. Scale bar, 100 μm. Quantification of the vessel length (h) and the number of branch points (i) in the cortical plate after coelectroporation. The data are represented as the means ± SEM (n = 3; one-way ANOVA; *P < 0.05). j Model showing how PRDM16 regulates angiogenesis through neuronal SMOC1 in the developing cortex. PRDM16 regulates SMOC1 secretion from neurons, and then neuronal SMOC1 activates the vascular TGF-β signaling pathway through interactions with the TGFBR1 receptor during brain development.