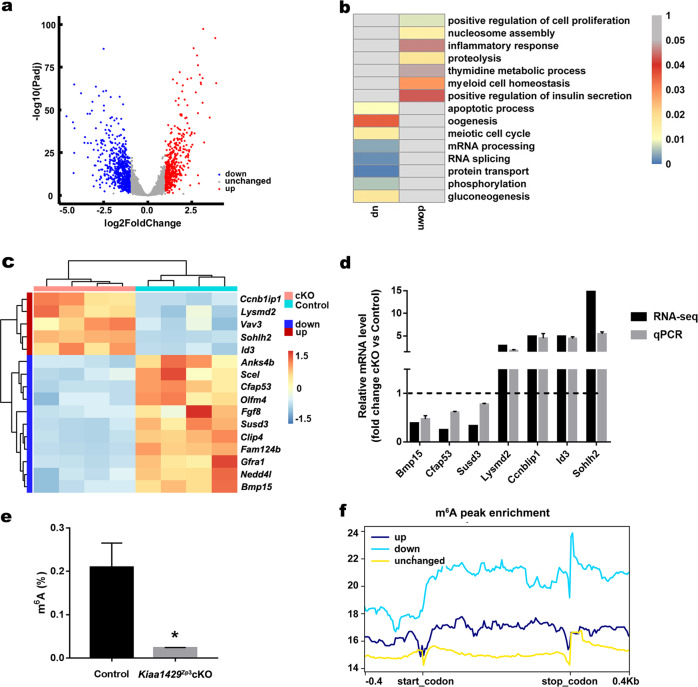
Fig. 4. Transcriptional profiling by RNA-seq and relevance to m6A modification.
a Volcano plots of the significantly differentially expressed genes (an adjusted p value < 0.05 and Fold change > 2; downregulated (blue), upregulated (Red), and unchanged genes (Gray) in Kiaa1429Zp3cKO oocytes are shown). b Enriched GO terms in downregulated and upregulated genes after Kiaa1429 deletion. The heatmap shows the −log10(p value) of the GO terms. c Heatmap of oocyte-derived factor genes. The color indicates the expression level after z-scoring. d Validation of RNA-seq data in oocytes from 20-day-old mice by qPCR. The tested genes were randomly selected from oocyte-derived factor genes. The RNA levels are shown relative to those of the control oocytes and were normalized to 1. Data of qPCR are presented as the mean ± s.e.m. from three independent experiments. e The m6A levels in total RNA from 20-day-old GV oocytes were measured using the EpiQuik m6A RNA Methylation Quantification Kit (EpiGentek). Data are presented as the mean ± s.e.m. from three independent experiments. *P < 0.05, t test (two-tailed). f The enrichment of m6A peaks in downregulated, upregulated, and unchanged genes after Kiaa1429 deletion.